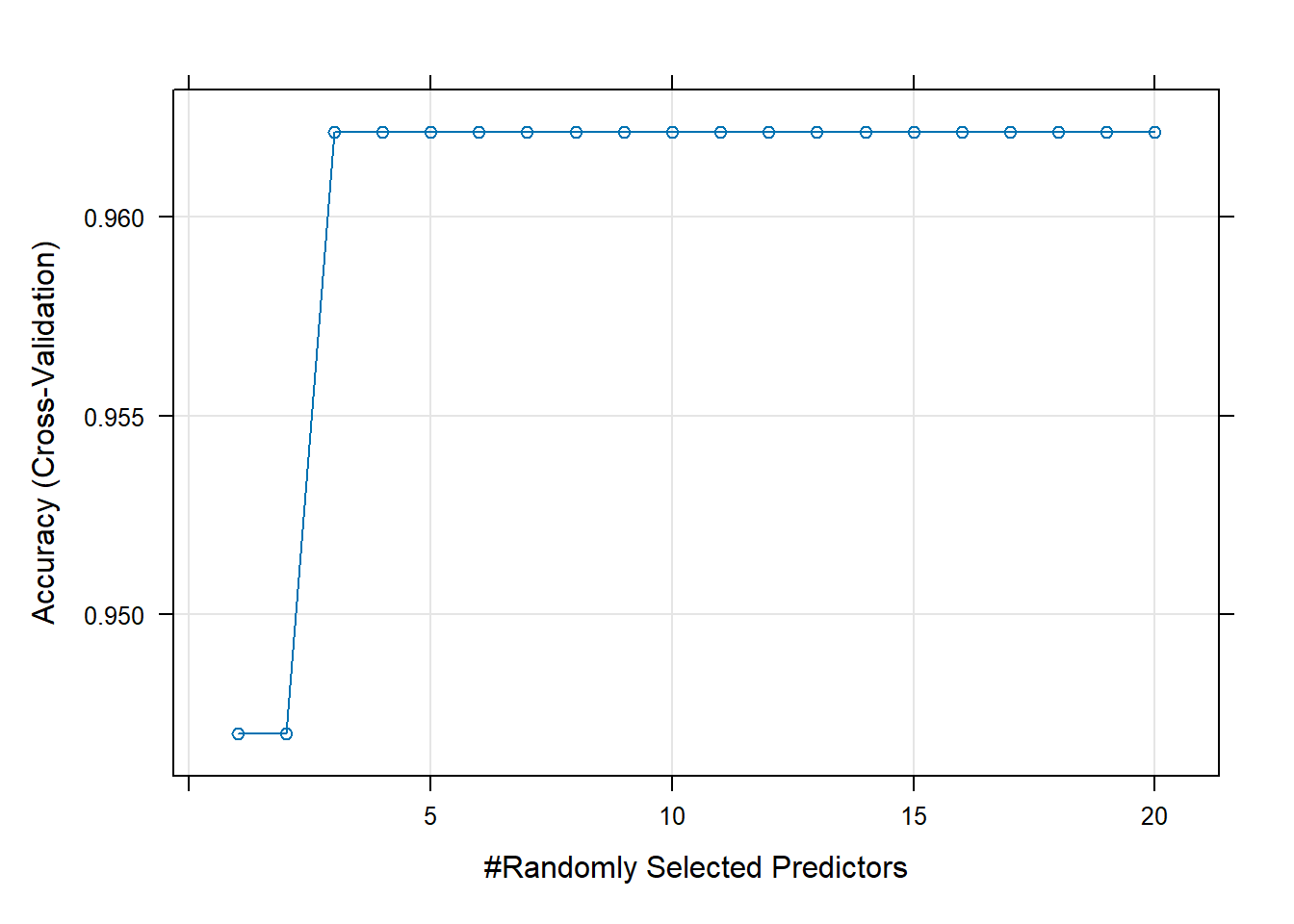
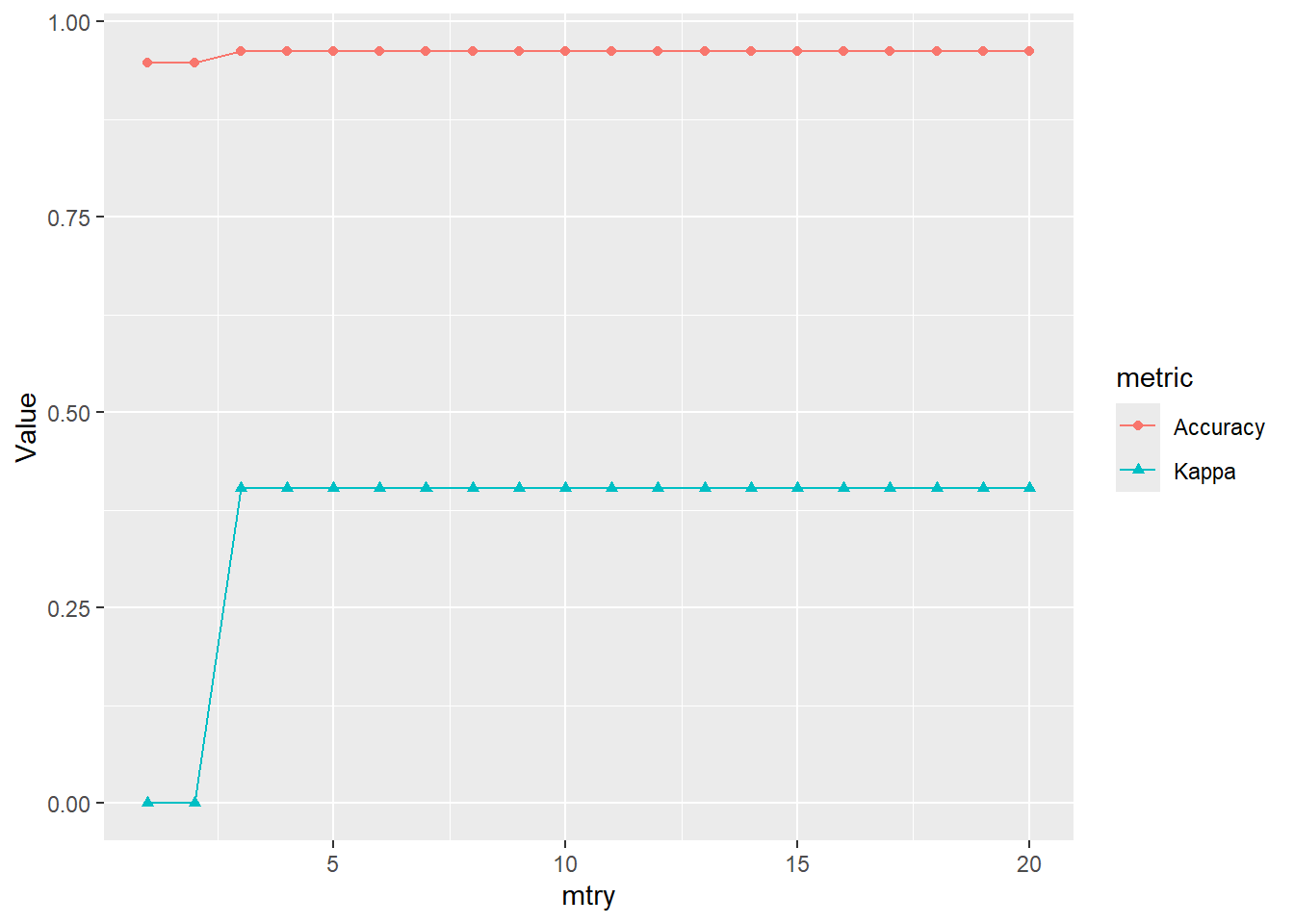
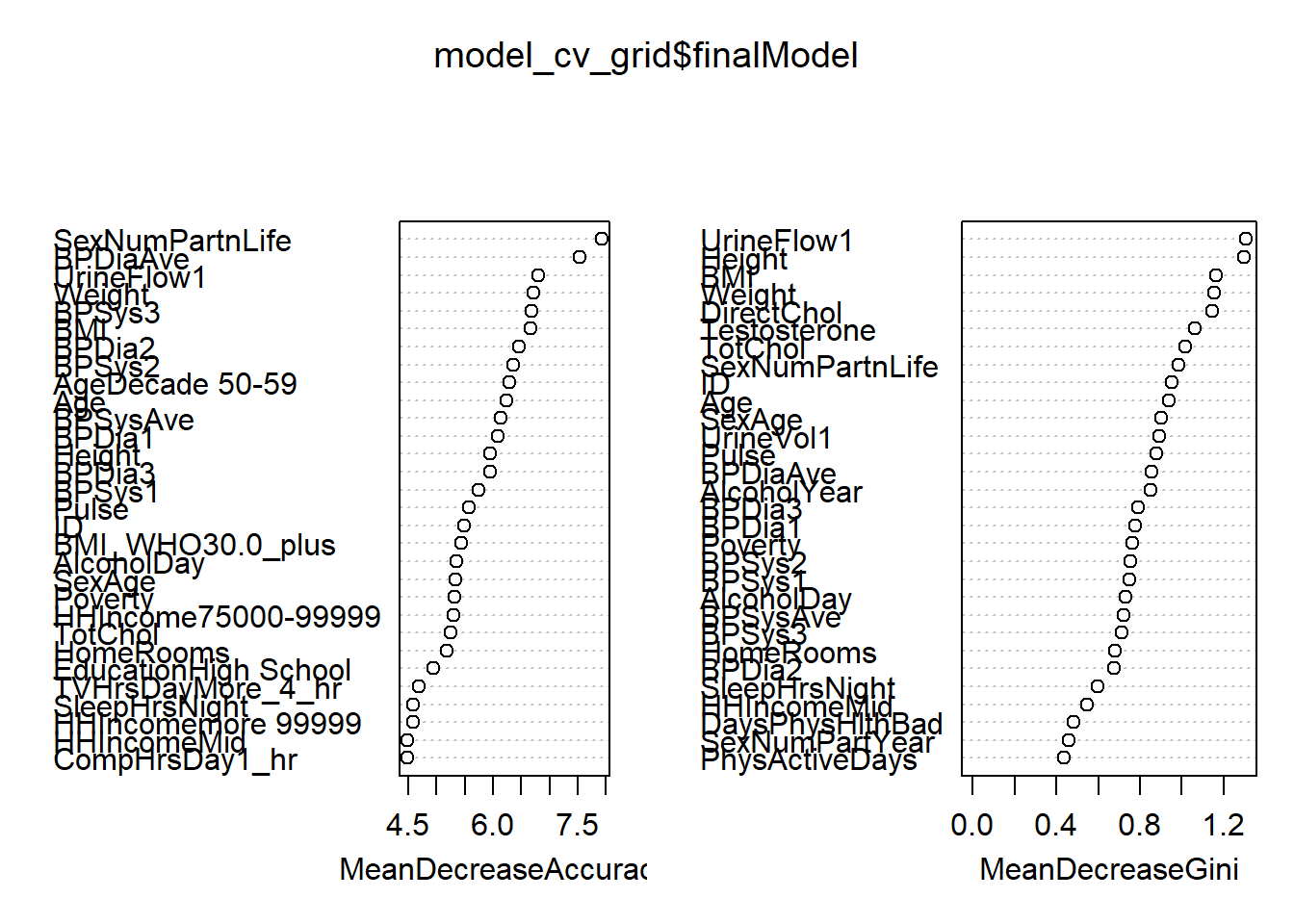
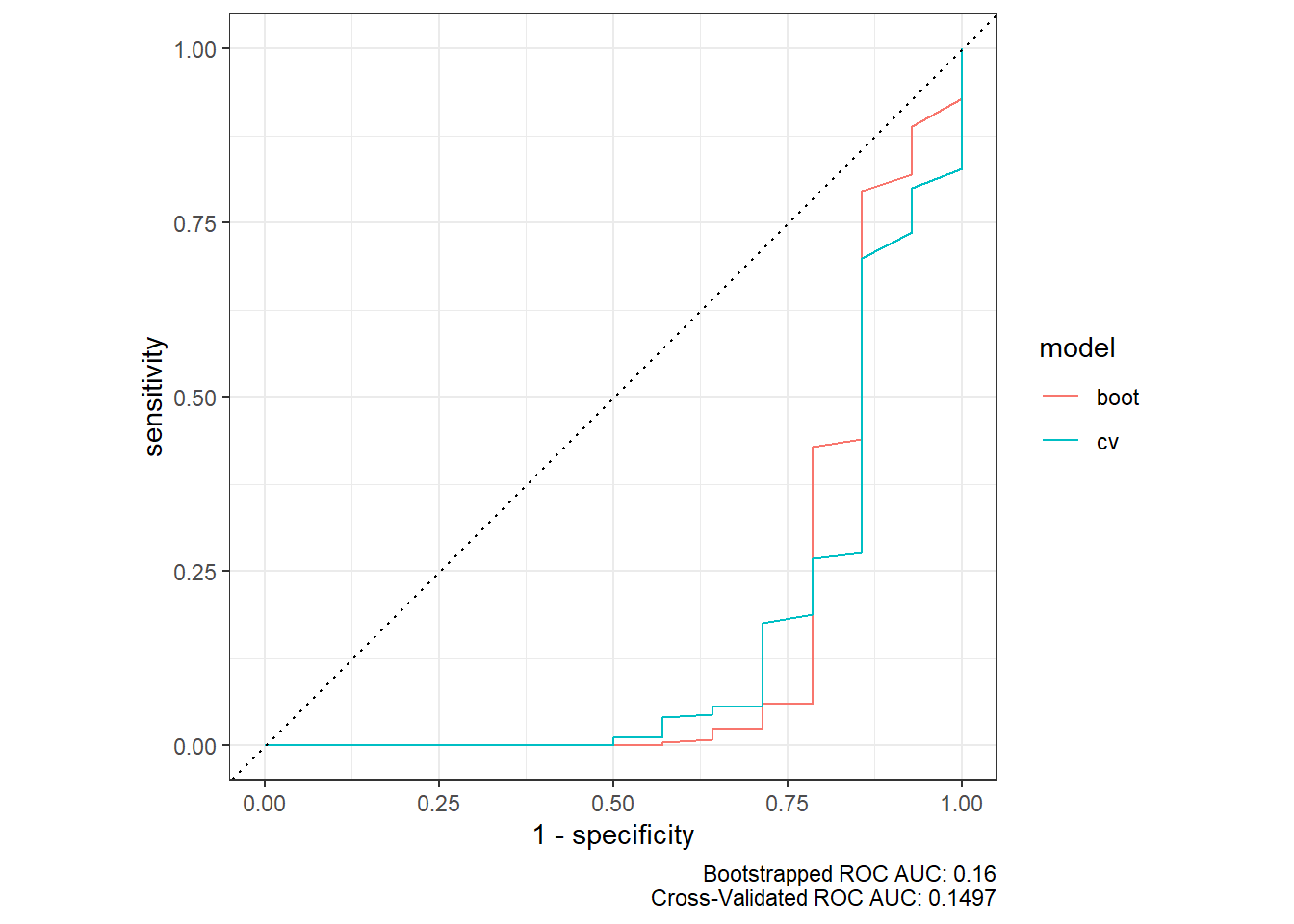
Rows: 264
Columns: 62
$ ID <int> 62172, 62199, 62231, 62340, 62444, 62460, 62481, 62552…
$ Diabetes <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No…
$ PhysActiveDays <int> 2, 7, 3, 5, 6, 3, 3, 7, 1, 1, 2, 1, 7, 3, 5, 4, 3, 3, …
$ SexOrientation <fct> Heterosexual, Homosexual, Heterosexual, Heterosexual, …
$ SexNumPartYear <int> 2, 0, 1, 1, 2, 1, 2, 1, 1, 2, 2, 10, 1, 1, 0, 1, 1, 1,…
$ Marijuana <fct> Yes, Yes, No, No, Yes, Yes, No, Yes, No, No, No, Yes, …
$ RegularMarij <fct> No, No, No, No, No, No, No, No, No, No, No, Yes, No, Y…
$ AlcoholDay <int> 3, 1, 2, 1, 4, 3, 2, 1, 1, 6, 6, 15, 1, 2, 1, 1, 1, 2,…
$ SexAge <int> 17, 19, 14, 19, 17, 16, 20, 16, 24, 17, 17, 17, 18, 14…
$ SexNumPartnLife <int> 4, 6, 1, 5, 3, 50, 25, 10, 7, 7, 7, 300, 4, 5, 5, 1, 1…
$ HardDrugs <fct> No, Yes, No, No, No, Yes, No, No, No, No, No, Yes, No,…
$ SexEver <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes,…
$ SameSex <fct> No, Yes, No, No, No, No, No, No, No, No, No, No, No, N…
$ AlcoholYear <int> 104, 260, 3, 12, 52, 104, 6, 300, 24, 52, 52, 104, 52,…
$ LittleInterest <fct> Several, None, None, Most, None, Several, None, None, …
$ Depressed <fct> Most, None, None, Most, None, Several, None, None, Non…
$ Alcohol12PlusYr <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes,…
$ Education <fct> High School, College Grad, High School, Some College, …
$ MaritalStatus <fct> NeverMarried, LivePartner, Married, Married, NeverMarr…
$ Smoke100 <fct> Yes, Yes, No, No, No, Yes, No, Yes, No, Yes, Yes, Yes,…
$ Smoke100n <fct> Smoker, Smoker, Non-Smoker, Non-Smoker, Non-Smoker, Sm…
$ DaysPhysHlthBad <int> 2, 0, 0, 0, 0, 0, 2, 0, 0, 1, 1, 7, 0, 2, 0, 0, 0, 0, …
$ DaysMentHlthBad <int> 10, 1, 0, 26, 0, 3, 30, 5, 0, 0, 0, 14, 0, 2, 0, 0, 0,…
$ HealthGen <fct> Good, Vgood, Good, Vgood, Fair, Good, Vgood, Vgood, Ex…
$ SleepHrsNight <int> 8, 8, 4, 6, 5, 6, 6, 8, 7, 6, 6, 6, 8, 6, 9, 7, 7, 6, …
$ Work <fct> NotWorking, Working, Working, Working, Working, Workin…
$ SleepTrouble <fct> No, No, No, No, No, Yes, No, No, No, No, No, Yes, No, …
$ BPSys1 <int> 100, 112, 120, 106, 112, 120, 98, 118, 102, 132, 132, …
$ BPDia1 <int> 70, 70, 70, 80, 54, 90, 56, 76, 62, 70, 70, 86, 76, 58…
$ Testosterone <dbl> 47.53, 269.24, 14.90, 296.66, 19.76, 299.19, 48.93, 18…
$ PhysActive <fct> No, Yes, No, No, Yes, Yes, No, Yes, Yes, Yes, Yes, Yes…
$ BPSys2 <int> 102, 108, 116, 106, 116, 124, 96, 108, 100, 124, 124, …
$ BPDia2 <int> 68, 64, 70, 82, 54, 88, 62, 80, 68, 66, 66, 72, 74, 62…
$ UrineFlow1 <dbl> 0.645, 0.380, 0.196, 0.623, 0.356, 0.498, 1.300, 1.500…
$ BPSys3 <int> 104, 112, 112, 106, 112, 130, 94, 120, 102, 126, 126, …
$ BPDia3 <int> 76, 66, 68, 80, 56, 90, 54, 76, 60, 70, 70, 70, 76, 60…
$ DirectChol <dbl> 1.89, 0.91, 1.53, 0.98, 1.16, 1.37, 1.19, 1.81, 1.47, …
$ TotChol <dbl> 4.37, 4.42, 4.71, 4.16, 4.34, 4.65, 3.83, 4.89, 4.34, …
$ BPSysAve <int> 103, 110, 114, 106, 114, 127, 95, 114, 101, 125, 125, …
$ BPDiaAve <int> 72, 65, 69, 81, 55, 89, 58, 78, 64, 68, 68, 71, 75, 61…
$ Pulse <int> 80, 84, 64, 68, 68, 68, 78, 84, 72, 80, 80, 78, 58, 60…
$ UrineVol1 <int> 107, 65, 19, 96, 26, 118, 282, 72, 276, 106, 106, 91, …
$ HHIncome <fct> 20000-24999, more 99999, more 99999, 25000-34999, 0-4…
$ HHIncomeMid <int> 22500, 100000, 100000, 30000, 2500, 100000, 70000, 100…
$ Poverty <dbl> 2.02, 5.00, 3.92, 1.28, 0.00, 4.34, 3.13, 4.07, 3.06, …
$ BMI_WHO <fct> 30.0_plus, 25.0_to_29.9, 30.0_plus, 25.0_to_29.9, 30.0…
$ AgeDecade <fct> 40-49, 50-59, 40-49, 40-49, 20-29, 40-49, 30-39…
$ BMI <dbl> 33.3, 28.0, 33.2, 25.9, 33.6, 21.9, 27.7, 22.5, 27.8, …
$ Height <dbl> 172.0, 186.0, 164.7, 173.2, 169.3, 188.1, 170.5, 160.8…
$ TVHrsDay <fct> More_4_hr, 1_hr, 2_hr, 3_hr, 4_hr, 1_hr, 1_hr, 2_hr, 0…
$ CompHrsDay <fct> More_4_hr, 1_hr, 0_to_1_hr, 1_hr, 1_hr, 0_to_1_hr, 3_h…
$ Weight <dbl> 98.6, 96.9, 90.1, 77.6, 96.4, 77.5, 80.5, 58.2, 87.2, …
$ HomeRooms <int> 4, 4, 6, 6, 5, 11, 5, 7, 10, 1, 1, 8, 8, 13, 5, 8, 8, …
$ HomeOwn <fct> Rent, Rent, Own, Own, Own, Own, Rent, Own, Own, Rent, …
$ SurveyYr <fct> 2011_12, 2011_12, 2011_12, 2011_12, 2011_12, 2011_12, …
$ Gender <fct> female, male, female, male, female, male, female, fema…
$ Age <int> 43, 57, 48, 44, 23, 41, 39, 41, 32, 21, 21, 35, 57, 29…
$ Race1 <fct> Black, White, Mexican, White, Black, White, Hispanic, …
$ Race3 <fct> Black, White, Mexican, White, Black, White, Hispanic, …
$ No <dbl> 0.9460189, 0.9716599, 0.9230769, 0.9919028, 0.9527665,…
$ Yes <dbl> 0.053981107, 0.028340081, 0.076923077, 0.008097166, 0.…
$ class <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, No…