9 k-Nearest Neighbors with caret
9.0.1 Reminders about the Data
tibble::tribble(
~"Variable in Data", ~"Definition", ~"Data Type",
'seqn', 'Respondent sequence number', 'Identifier',
'riagendr', 'Gender', 'Categorical',
'ridageyr', 'Age in years at screening', 'Continuous / Numerical',
'ridreth1', 'Race/Hispanic origin', 'Categorical',
'dmdeduc2', 'Education level', 'Adults 20+ - Categorical',
'dmdmartl', 'Marital status', 'Categorical',
'indhhin2', 'Annual household income', 'Categorical',
'bmxbmi', 'Body Mass Index (kg/m**2)', 'Continuous / Numerical',
'diq010', 'Doctor diagnosed diabetes', 'Categorical',
'lbxglu', 'Fasting Glucose (mg/dL)', 'Continuous / Numerical'
) |>
knitr::kable()| Variable in Data | Definition | Data Type |
|---|---|---|
| seqn | Respondent sequence number | Identifier |
| riagendr | Gender | Categorical |
| ridageyr | Age in years at screening | Continuous / Numerical |
| ridreth1 | Race/Hispanic origin | Categorical |
| dmdeduc2 | Education level | Adults 20+ - Categorical |
| dmdmartl | Marital status | Categorical |
| indhhin2 | Annual household income | Categorical |
| bmxbmi | Body Mass Index (kg/m**2) | Continuous / Numerical |
| diq010 | Doctor diagnosed diabetes | Categorical |
| lbxglu | Fasting Glucose (mg/dL) | Continuous / Numerical |
9.0.2 Install if not Function
install_if_not <- function( list.of.packages ) {
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) { install.packages(new.packages) } else { print(paste0("the package '", list.of.packages , "' is already installed")) }
}
9.1 Load tidyverse
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
9.2 The caret package
9.3 Split Data
# The createDataPartition function is used to create training and test sets
trainIndex <- createDataPartition(diab_pop.no_na_vals$diq010,
p = .6,
list = FALSE,
times = 1)
dm2.train <- diab_pop.no_na_vals[trainIndex, ]
dm2.test <- diab_pop.no_na_vals[-trainIndex, ]9.4 Make Grid
# we will make a grid of values to test in cross-validation.
knnGrid <- expand.grid(k = 1:15)9.4.1 Optimize for Accuracy
9.4.1.1 The trainControl function
# the method here is cv for cross-validation you could try "repeatedcv" for repeated cross-fold validation
fitControl <- trainControl(method = "cv", # uncomment for repeatedcv
number = 10,
# repeats = 10, # uncomment for repeatedcv
## Estimate class probabilities
classProbs = TRUE)
9.4.1.2 The train function
knnFit <- train(diq010 ~ ., # formula
data = dm2.train, # train data
method = "knn", # method for caret see https://topepo.github.io/caret/available-models.html for list of models
trControl = fitControl,
tuneGrid = knnGrid,
metric = "Accuracy") ## Specify which metric to optimize9.4.1.3 Results
knnFitk-Nearest Neighbors
1126 samples
9 predictor
2 classes: 'Diabetes', 'No_Diabetes'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 1013, 1013, 1013, 1014, 1013, 1013, ...
Resampling results across tuning parameters:
k Accuracy Kappa
1 0.8552070 0.38286631
2 0.8471871 0.32674490
3 0.8693979 0.32082482
4 0.8649652 0.25745853
5 0.8703303 0.24645605
6 0.8614728 0.17410430
7 0.8614570 0.12685254
8 0.8641277 0.15354040
9 0.8605563 0.11030512
10 0.8570006 0.07271797
11 0.8561157 0.06507766
12 0.8543458 0.04587766
13 0.8525759 0.02878628
14 0.8507980 0.00960000
15 0.8516909 0.01985641
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was k = 5.plot(knnFit)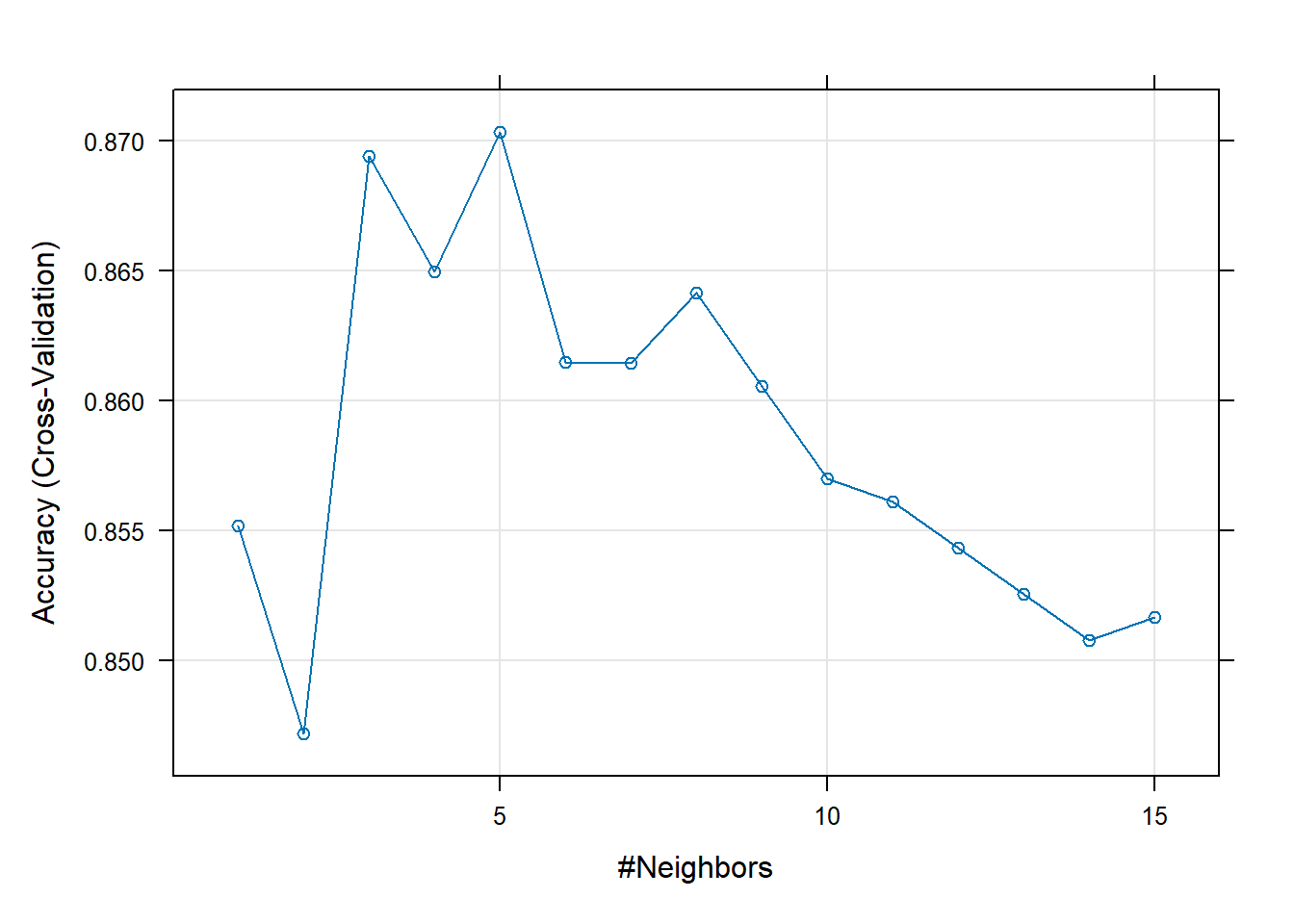
str(knnFit,1)List of 25
$ method : chr "knn"
$ modelInfo :List of 13
$ modelType : chr "Classification"
$ results :'data.frame': 15 obs. of 5 variables:
$ pred : NULL
$ bestTune :'data.frame': 1 obs. of 1 variable:
$ call : language train.formula(form = diq010 ~ ., data = dm2.train, method = "knn", trControl = fitControl, tuneGrid = knnGri| __truncated__
$ dots : list()
$ metric : chr "Accuracy"
$ control :List of 27
$ finalModel :List of 8
..- attr(*, "class")= chr "knn3"
$ preProcess : NULL
$ trainingData:'data.frame': 1126 obs. of 10 variables:
$ ptype :'data.frame': 0 obs. of 9 variables:
$ resample :'data.frame': 10 obs. of 3 variables:
$ resampledCM :'data.frame': 150 obs. of 6 variables:
$ perfNames : chr [1:2] "Accuracy" "Kappa"
$ maximize : logi TRUE
$ yLimits : NULL
$ times :List of 3
$ levels : chr [1:2] "Diabetes" "No_Diabetes"
..- attr(*, "ordered")= logi FALSE
$ terms :Classes 'terms', 'formula' language diq010 ~ seqn + riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu
.. ..- attr(*, "variables")= language list(diq010, seqn, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "factors")= int [1:10, 1:9] 0 1 0 0 0 0 0 0 0 0 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "term.labels")= chr [1:9] "seqn" "riagendr" "ridageyr" "ridreth1" ...
.. ..- attr(*, "order")= int [1:9] 1 1 1 1 1 1 1 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(diq010, seqn, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "dataClasses")= Named chr [1:10] "factor" "numeric" "factor" "numeric" ...
.. .. ..- attr(*, "names")= chr [1:10] "diq010" "seqn" "riagendr" "ridageyr" ...
$ coefnames : chr [1:31] "seqn" "riagendrFemale" "ridageyr" "ridreth1Other Hispanic" ...
$ contrasts :List of 5
$ xlevels :List of 5
- attr(*, "class")= chr [1:2] "train" "train.formula"knnFit$finalModel 5-nearest neighbor model
Training set outcome distribution:
Diabetes No_Diabetes
169 957 9.4.1.3.1 Score Test Data
# let's score the test set using this model
pred_class <- predict(knnFit, dm2.test,'raw')
probs <- predict(knnFit, dm2.test,'prob')
dm2.test.scored <- cbind(dm2.test, pred_class, probs)
glimpse(dm2.test.scored)Rows: 750
Columns: 13
$ seqn <dbl> 83734, 83737, 83757, 83761, 83789, 83820, 83822, 83823, 83…
$ riagendr <fct> Male, Female, Female, Female, Male, Male, Female, Female, …
$ ridageyr <dbl> 78, 72, 57, 24, 66, 70, 20, 29, 69, 71, 37, 49, 41, 54, 80…
$ ridreth1 <fct> Non-Hispanic White, MexicanAmerican, Other Hispanic, Other…
$ dmdeduc2 <fct> High school graduate/GED, Grades 9-11th, Less than 9th gra…
$ dmdmartl <fct> Married, Separated, Separated, Never married, Living with …
$ indhhin2 <fct> "$20,000-$24,999", "$75,000-$99,999", "$20,000-$24,999", "…
$ bmxbmi <dbl> 28.8, 28.6, 35.4, 25.3, 34.0, 27.0, 22.2, 29.7, 28.2, 27.6…
$ diq010 <fct> Diabetes, No_Diabetes, Diabetes, No_Diabetes, No_Diabetes,…
$ lbxglu <dbl> 84, 107, 398, 95, 113, 94, 80, 102, 105, 76, 79, 126, 110,…
$ pred_class <fct> No_Diabetes, No_Diabetes, No_Diabetes, No_Diabetes, No_Dia…
$ Diabetes <dbl> 0.0, 0.0, 0.4, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2, 0.0, 0.2…
$ No_Diabetes <dbl> 1.0, 1.0, 0.6, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 0.8, 1.0, 0.8…
9.4.1.3.2 Use yardstick for model metrics
# yardstick
library('yardstick')
Attaching package: 'yardstick'The following objects are masked from 'package:caret':
precision, recall, sensitivity, specificityThe following object is masked from 'package:readr':
spec9.4.1.3.3 Confusion Matrix
Truth
Prediction Diabetes No_Diabetes
Diabetes 14 3
No_Diabetes 98 635# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.865
2 kap binary 0.185
3 sens binary 0.125
4 spec binary 0.995
5 ppv binary 0.824
6 npv binary 0.866
7 mcc binary 0.288
8 j_index binary 0.120
9 bal_accuracy binary 0.560
10 detection_prevalence binary 0.0227
11 precision binary 0.824
12 recall binary 0.125
13 f_meas binary 0.217 dm2.test.scored %>%
conf_mat(truth=diq010 , pred_class) %>%
summary() %>%
ggplot(aes(y=.metric, x=.estimate, fill=.metric)) +
geom_bar(stat="identity")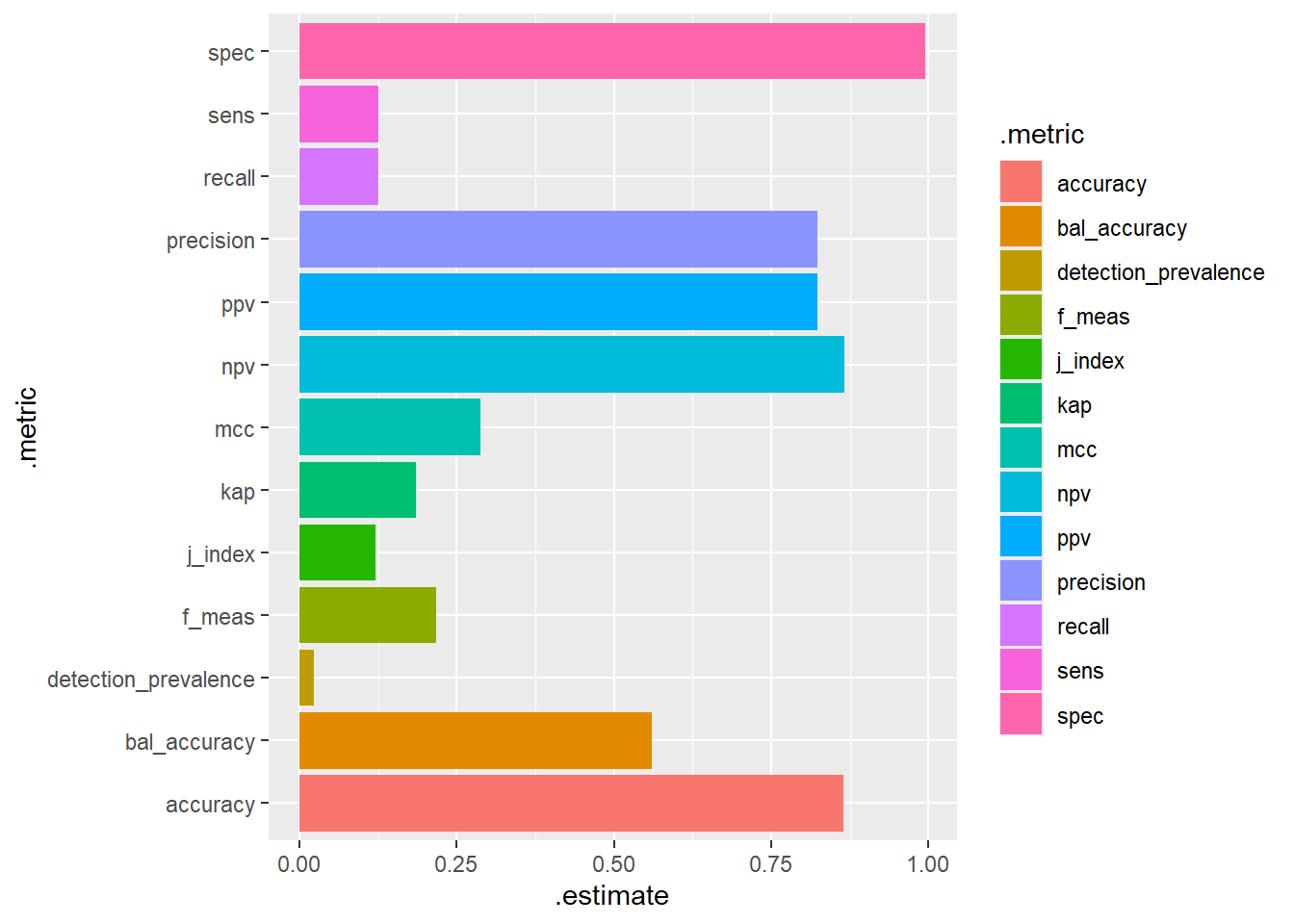
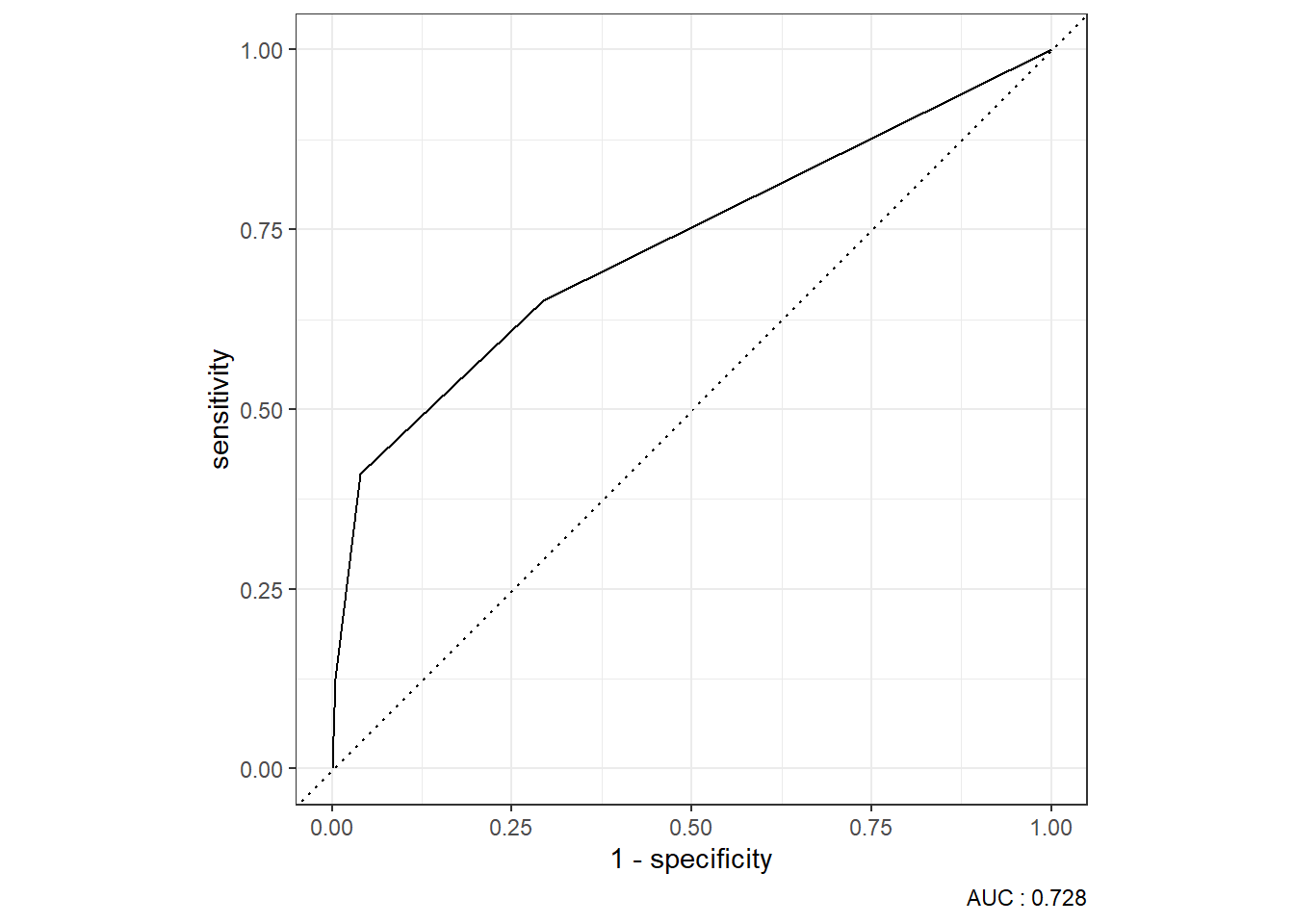
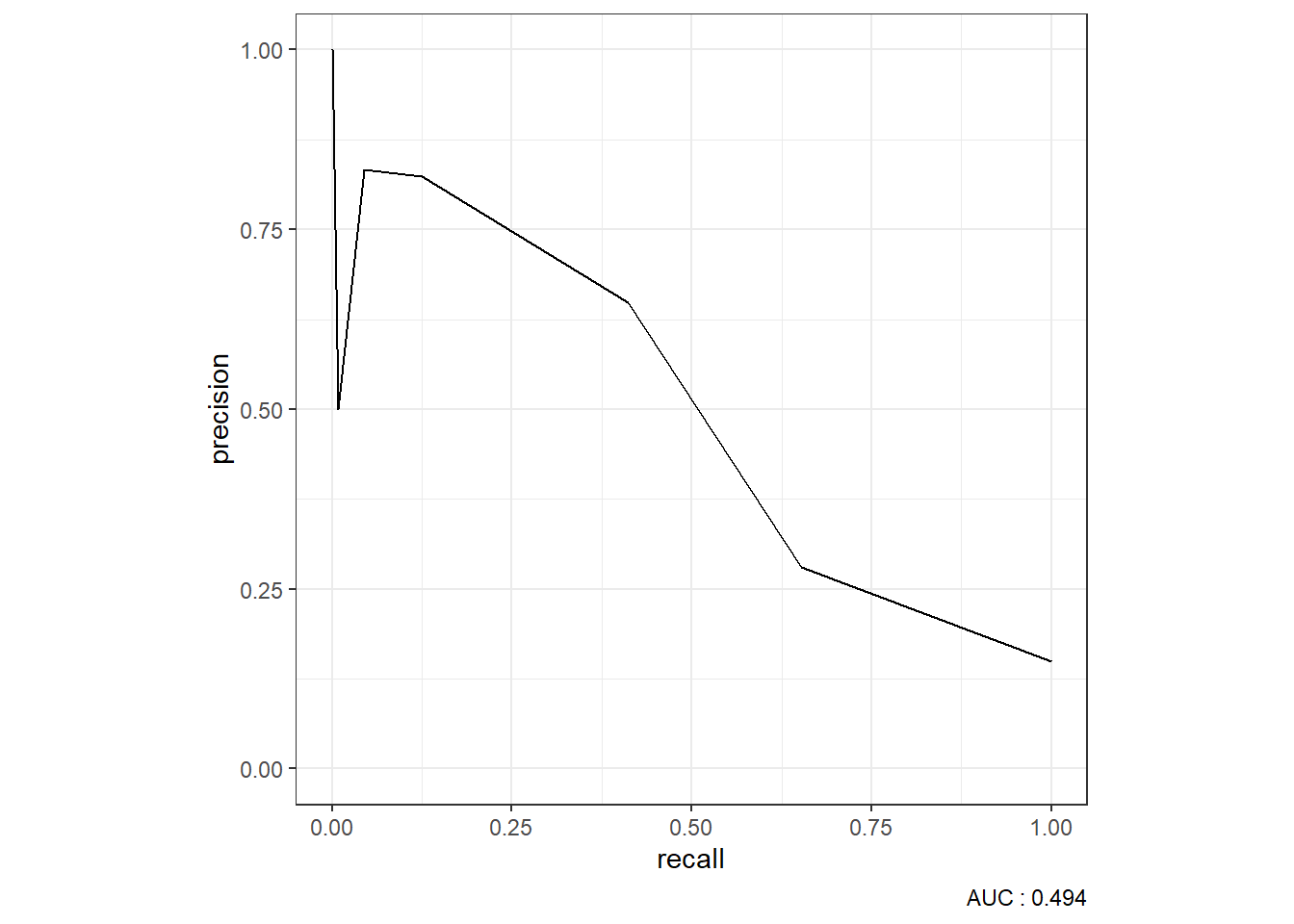
9.4.1.3.4 ROC Curve
9.4.1.3.5 Precision-Recall Curve
9.4.2 Optimize for ROC
9.4.2.1 The trainControl function
# the method here is cv for cross-validation you could try "repeatedcv" for repeated cross-fold validation
fitControl <- trainControl(method = "cv", # uncomment for repeatedcv
number = 10,
# repeats = 10, # uncomment for repeatedcv
## Estimate class probabilities
classProbs = TRUE,
summaryFunction=twoClassSummary)
9.4.2.2 The train function
knnFit_ROC <- train(diq010 ~ ., # formula
data = dm2.train, # train data
method = "knn", # method for caret see https://topepo.github.io/caret/available-models.html for list of models
trControl = fitControl,
tuneGrid = knnGrid,
metric = "ROC") ## Specify which metric to optimize9.4.2.3 Results
knnFit_ROCk-Nearest Neighbors
1126 samples
9 predictor
2 classes: 'Diabetes', 'No_Diabetes'
No pre-processing
Resampling: Cross-Validated (10 fold)
Summary of sample sizes: 1013, 1014, 1014, 1014, 1013, 1013, ...
Resampling results across tuning parameters:
k ROC Sens Spec
1 0.6683833 0.395220588 0.9415461
2 0.7051856 0.389705882 0.9404276
3 0.7176461 0.265441176 0.9749342
4 0.7156281 0.224632353 0.9812061
5 0.7006004 0.153308824 0.9937281
6 0.6966088 0.135294118 0.9937390
7 0.6899122 0.076470588 0.9979167
8 0.6780983 0.076470588 0.9968750
9 0.6762613 0.058823529 1.0000000
10 0.6752873 0.052941176 0.9989583
11 0.6677884 0.035294118 1.0000000
12 0.6583659 0.023529412 1.0000000
13 0.6547260 0.011764706 1.0000000
14 0.6528311 0.005882353 1.0000000
15 0.6497179 0.011764706 1.0000000
ROC was used to select the optimal model using the largest value.
The final value used for the model was k = 3.plot(knnFit_ROC)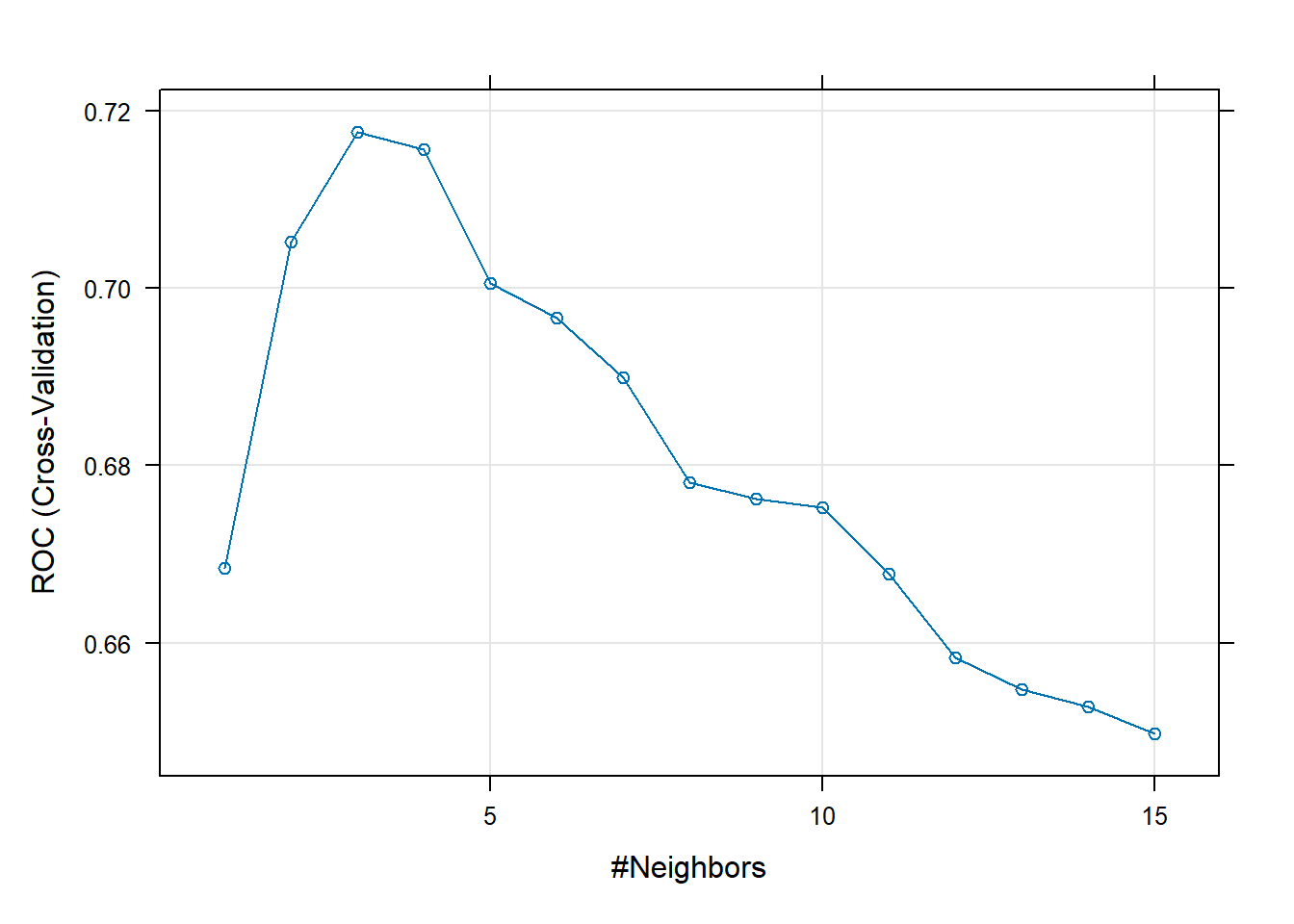
str(knnFit_ROC,1)List of 25
$ method : chr "knn"
$ modelInfo :List of 13
$ modelType : chr "Classification"
$ results :'data.frame': 15 obs. of 7 variables:
$ pred : NULL
$ bestTune :'data.frame': 1 obs. of 1 variable:
$ call : language train.formula(form = diq010 ~ ., data = dm2.train, method = "knn", trControl = fitControl, tuneGrid = knnGri| __truncated__
$ dots : list()
$ metric : chr "ROC"
$ control :List of 27
$ finalModel :List of 8
..- attr(*, "class")= chr "knn3"
$ preProcess : NULL
$ trainingData:'data.frame': 1126 obs. of 10 variables:
$ ptype :'data.frame': 0 obs. of 9 variables:
$ resample :'data.frame': 10 obs. of 4 variables:
$ resampledCM :'data.frame': 150 obs. of 6 variables:
$ perfNames : chr [1:3] "ROC" "Sens" "Spec"
$ maximize : logi TRUE
$ yLimits : NULL
$ times :List of 3
$ levels : chr [1:2] "Diabetes" "No_Diabetes"
..- attr(*, "ordered")= logi FALSE
$ terms :Classes 'terms', 'formula' language diq010 ~ seqn + riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu
.. ..- attr(*, "variables")= language list(diq010, seqn, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "factors")= int [1:10, 1:9] 0 1 0 0 0 0 0 0 0 0 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "term.labels")= chr [1:9] "seqn" "riagendr" "ridageyr" "ridreth1" ...
.. ..- attr(*, "order")= int [1:9] 1 1 1 1 1 1 1 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(diq010, seqn, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "dataClasses")= Named chr [1:10] "factor" "numeric" "factor" "numeric" ...
.. .. ..- attr(*, "names")= chr [1:10] "diq010" "seqn" "riagendr" "ridageyr" ...
$ coefnames : chr [1:31] "seqn" "riagendrFemale" "ridageyr" "ridreth1Other Hispanic" ...
$ contrasts :List of 5
$ xlevels :List of 5
- attr(*, "class")= chr [1:2] "train" "train.formula"knnFit_ROC$finalModel 3-nearest neighbor model
Training set outcome distribution:
Diabetes No_Diabetes
169 957 9.4.2.3.1 Score Test Data
# let's score the test set using this model
pred_class <- predict(knnFit_ROC, dm2.test, 'raw')
probs <- predict(knnFit_ROC, dm2.test, 'prob')
dm2.test.scored_ROC <- cbind(dm2.test, pred_class, probs)
glimpse(dm2.test.scored_ROC)Rows: 750
Columns: 13
$ seqn <dbl> 83734, 83737, 83757, 83761, 83789, 83820, 83822, 83823, 83…
$ riagendr <fct> Male, Female, Female, Female, Male, Male, Female, Female, …
$ ridageyr <dbl> 78, 72, 57, 24, 66, 70, 20, 29, 69, 71, 37, 49, 41, 54, 80…
$ ridreth1 <fct> Non-Hispanic White, MexicanAmerican, Other Hispanic, Other…
$ dmdeduc2 <fct> High school graduate/GED, Grades 9-11th, Less than 9th gra…
$ dmdmartl <fct> Married, Separated, Separated, Never married, Living with …
$ indhhin2 <fct> "$20,000-$24,999", "$75,000-$99,999", "$20,000-$24,999", "…
$ bmxbmi <dbl> 28.8, 28.6, 35.4, 25.3, 34.0, 27.0, 22.2, 29.7, 28.2, 27.6…
$ diq010 <fct> Diabetes, No_Diabetes, Diabetes, No_Diabetes, No_Diabetes,…
$ lbxglu <dbl> 84, 107, 398, 95, 113, 94, 80, 102, 105, 76, 79, 126, 110,…
$ pred_class <fct> No_Diabetes, No_Diabetes, Diabetes, No_Diabetes, No_Diabet…
$ Diabetes <dbl> 0.0000000, 0.0000000, 0.6666667, 0.0000000, 0.0000000, 0.0…
$ No_Diabetes <dbl> 1.0000000, 1.0000000, 0.3333333, 1.0000000, 1.0000000, 1.0…
9.4.2.3.2 Use yardstick for model metrics
9.4.2.3.3 Confusion Matrix
Truth
Prediction Diabetes No_Diabetes
Diabetes 31 9
No_Diabetes 81 629# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.88
2 kap binary 0.357
3 sens binary 0.277
4 spec binary 0.986
5 ppv binary 0.775
6 npv binary 0.886
7 mcc binary 0.417
8 j_index binary 0.263
9 bal_accuracy binary 0.631
10 detection_prevalence binary 0.0533
11 precision binary 0.775
12 recall binary 0.277
13 f_meas binary 0.408 dm2.test.scored_ROC %>%
conf_mat(truth=diq010 , pred_class) %>%
summary() %>%
ggplot(aes(y=.metric, x=.estimate, fill=.metric)) +
geom_bar(stat="identity")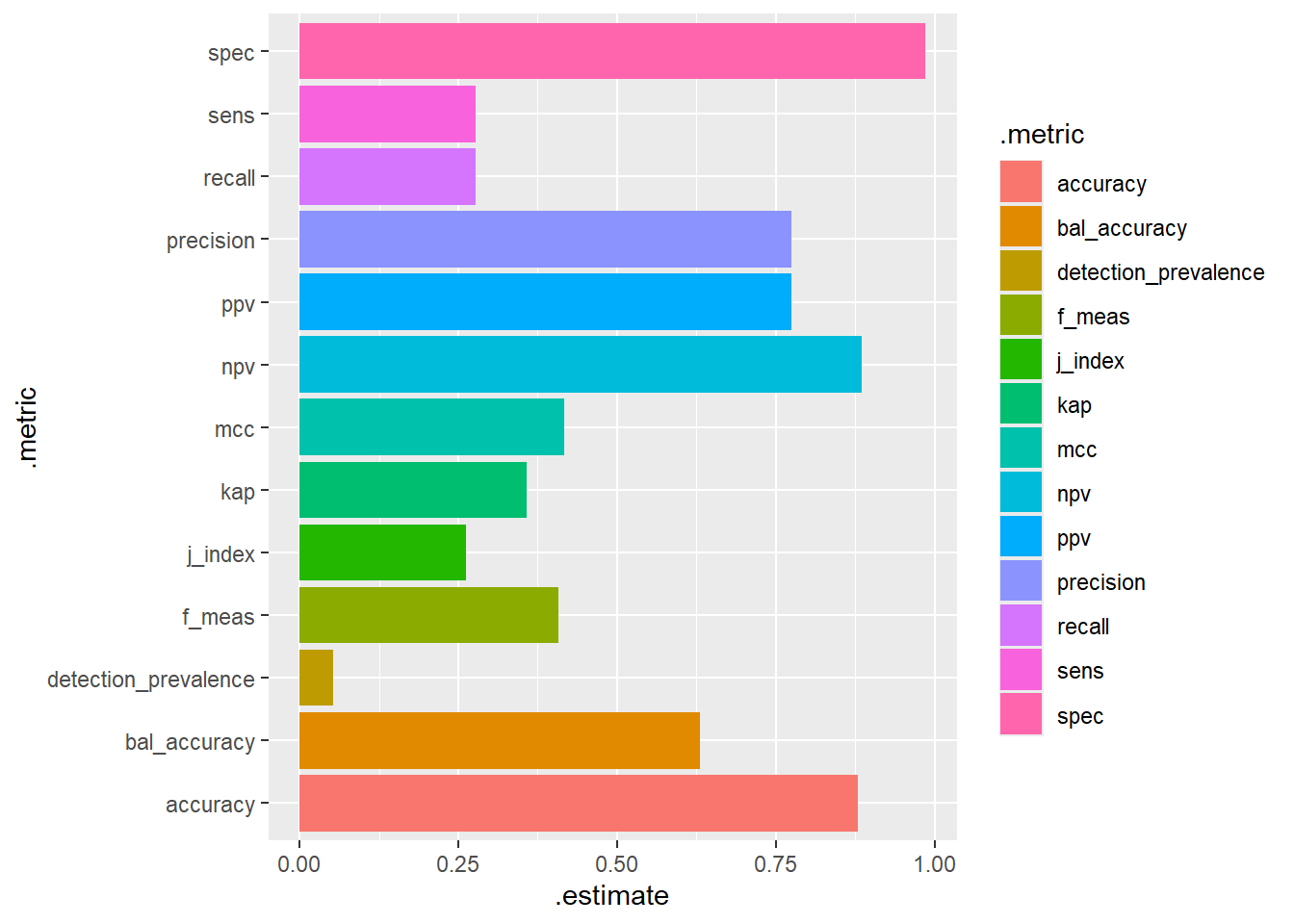
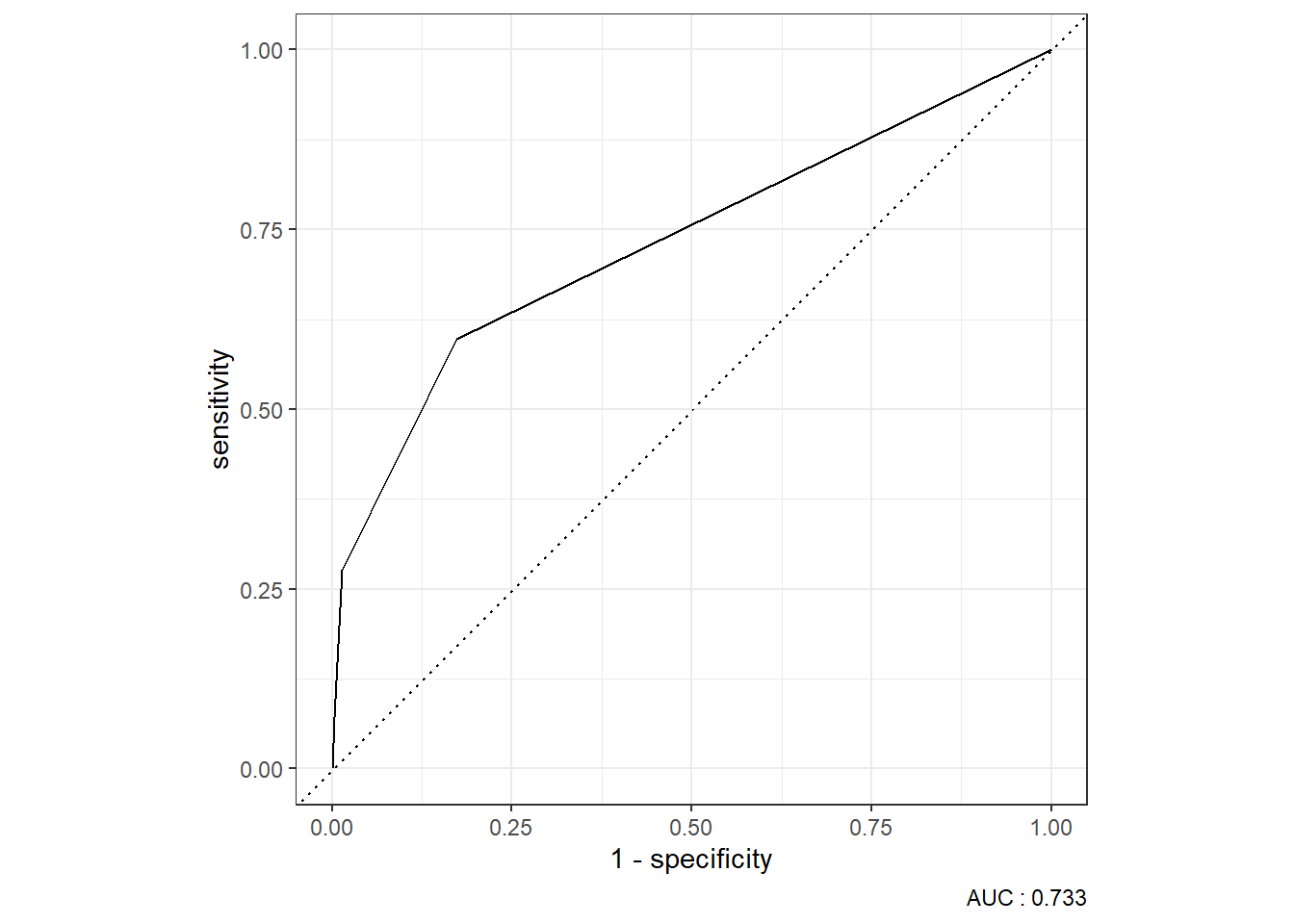
9.4.2.3.4 ROC Curve
9.4.2.3.5 Precision-Recall Curve
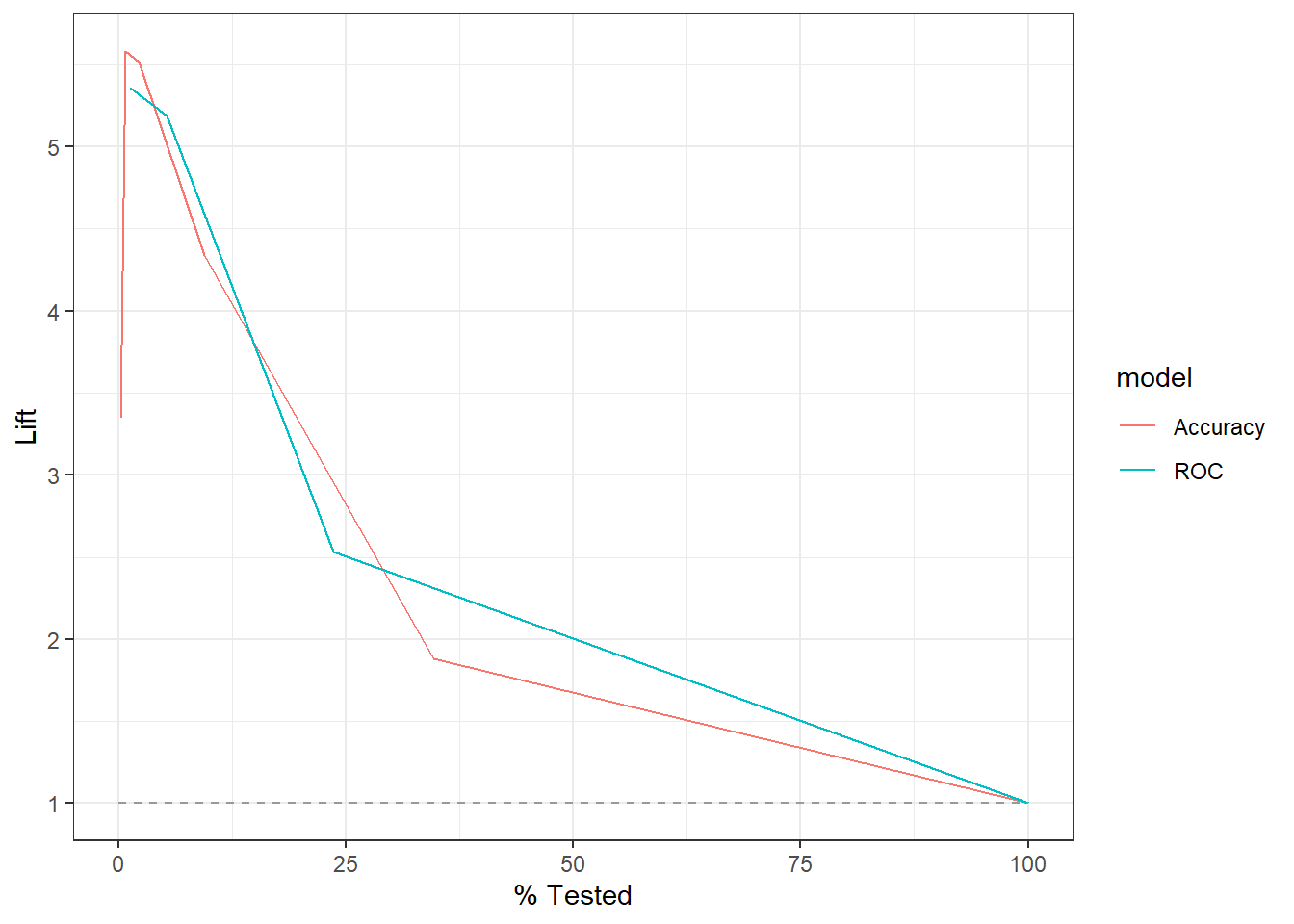
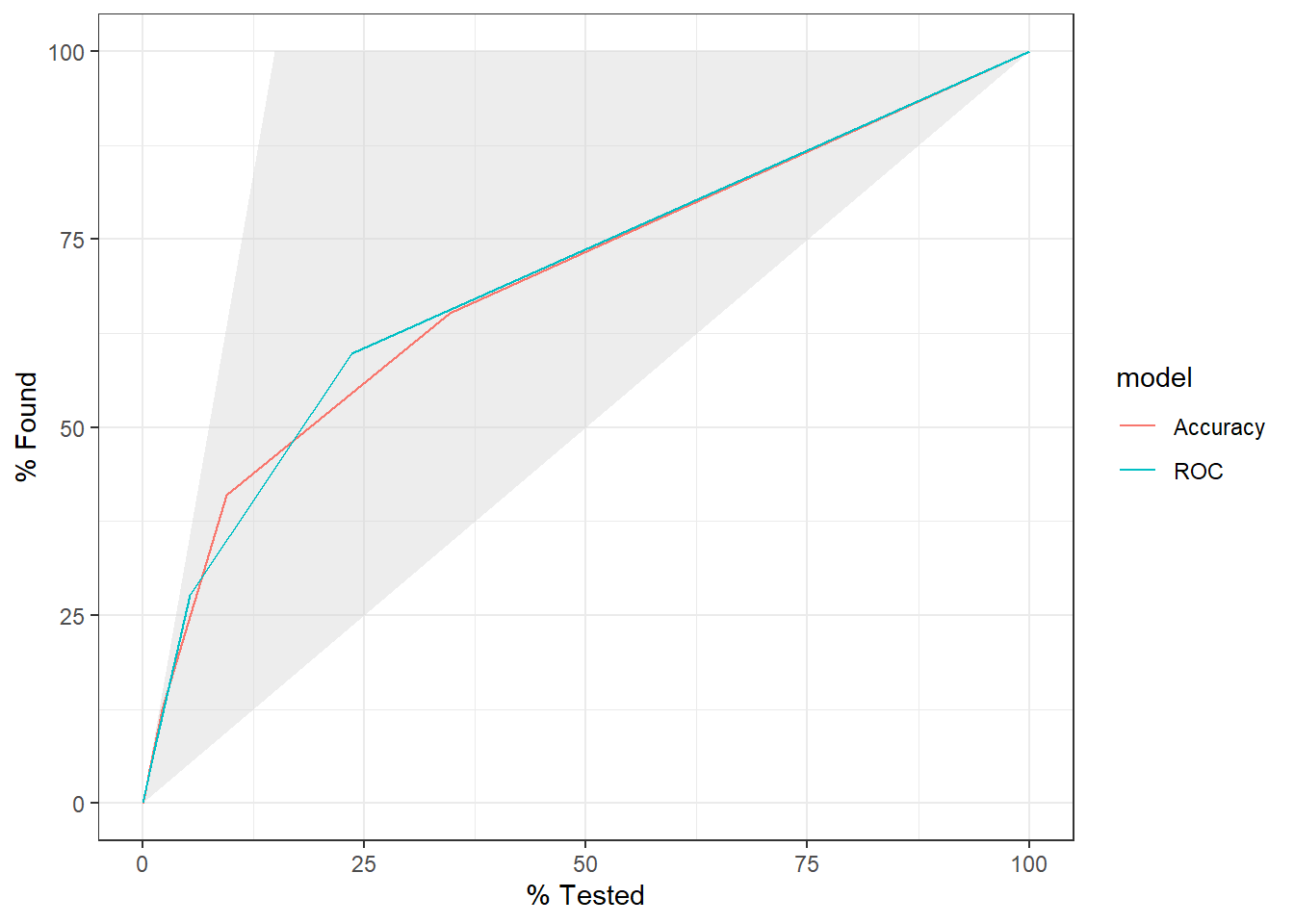
9.5 Compare Results
9.5.1 Confusion Matrix
knn_Fit_compare %>%
group_by(model) %>%
conf_mat(truth=diq010 , pred_class) %>%
ungroup() %>%
pull(conf_mat) %>%
map(summary) %>%
bind_rows(.id = "model") %>%
mutate(model = if_else(model == 1, "Accuracy", "ROC")) %>%
ggplot(aes(y=.metric, x=.estimate, fill=model, color = .metric)) +
geom_bar(stat="identity", position = 'dodge')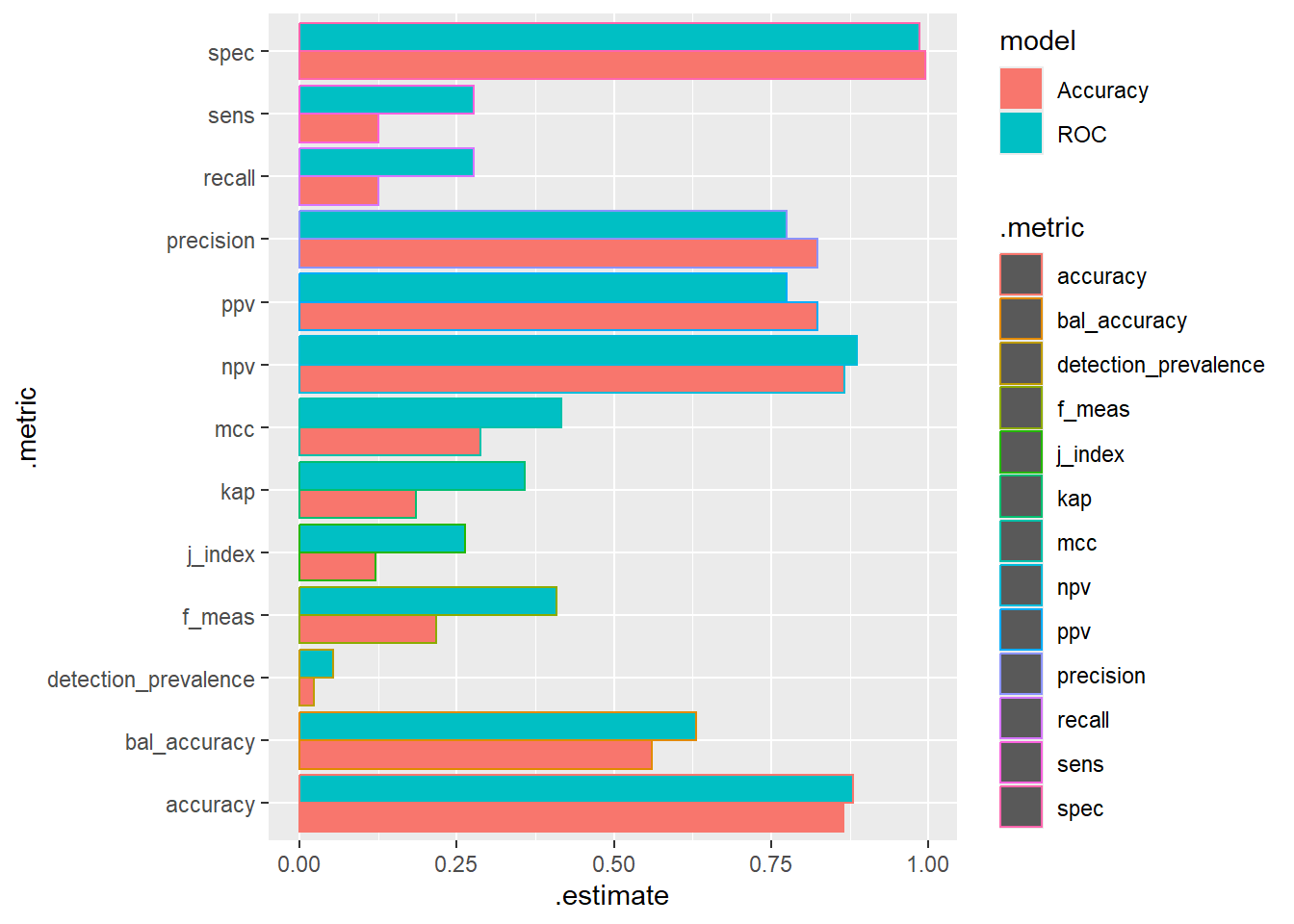
9.5.2 ROC Curve
knn_Fit_compare_AUC <- knn_Fit_compare %>%
group_by(model) %>%
roc_auc(truth=diq010 , Diabetes) %>%
mutate(AUC = paste(model, " AUC: ", round(.estimate,3))) %>%
ungroup()
knn_Fit_compare %>%
left_join(knn_Fit_compare_AUC) %>%
group_by(AUC) %>%
roc_curve(truth=diq010 , Diabetes) %>%
autoplot()Joining with `by = join_by(model)`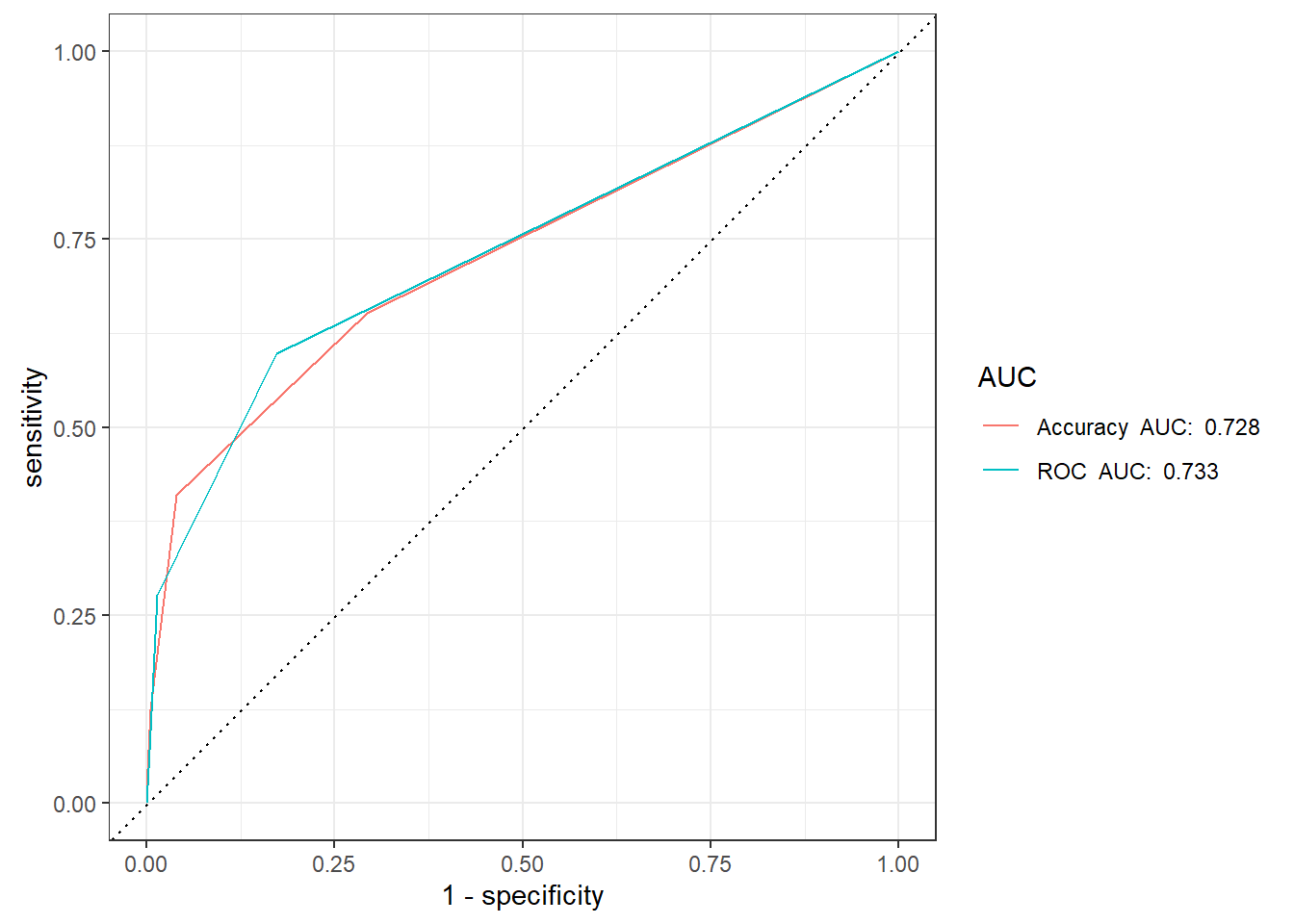
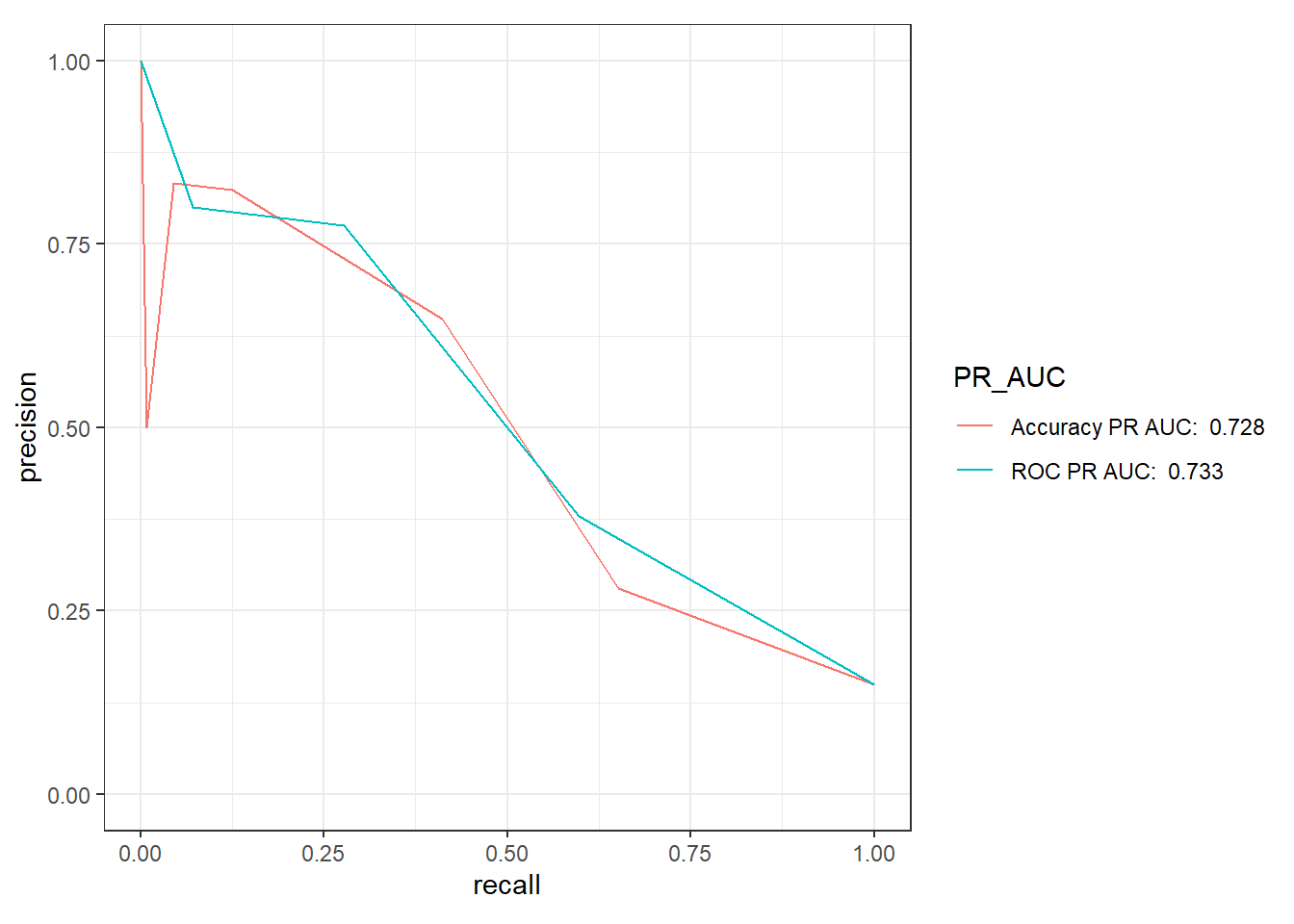
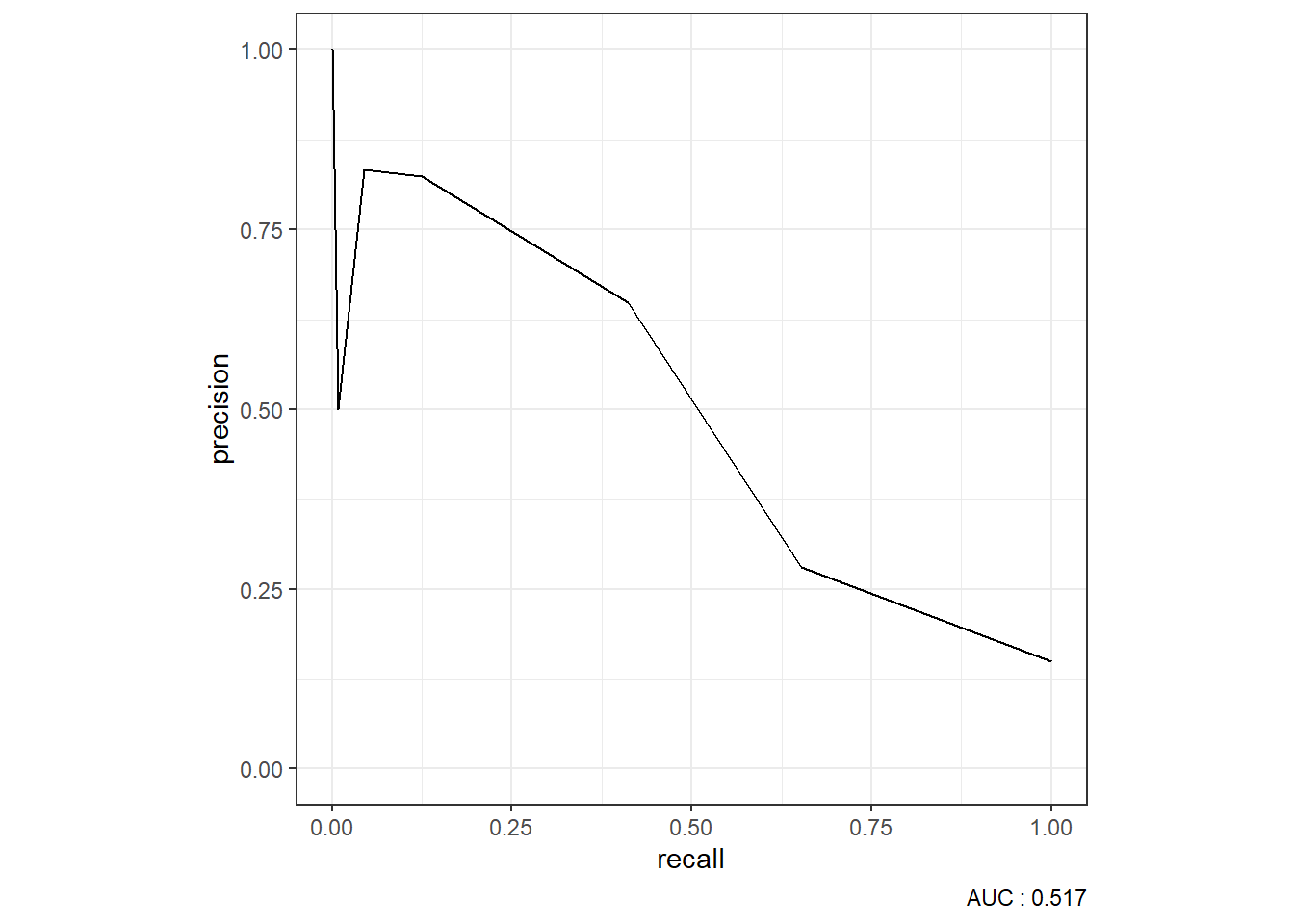
9.5.3 Precision-Recall Curve
knn_Fit_compare_PR <- knn_Fit_compare %>%
group_by(model) %>%
roc_auc(truth=diq010 , Diabetes) %>%
mutate(PR_AUC = paste(model, "PR AUC: ", round(.estimate,3))) %>%
ungroup()
knn_Fit_compare %>%
left_join(knn_Fit_compare_PR) %>%
group_by(PR_AUC) %>%
pr_curve(truth=diq010 , Diabetes) %>%
autoplot()Joining with `by = join_by(model)`