diab_pop <- readRDS('C:/Users/jkyle/Documents/GitHub/Intro_Jeff_Data_Science/DATA/diab_pop.RDS')21 Read in the Data
#Decision Trees
\(~\)
\(~\)
\(~\)
\(~\)
\(~\)
\(~\)
21.1 Reminders
###The Data
#### Variable in Data - Definition - Data Type
##### seqn - Respondent sequence number - Identifier
##### riagendr - Gender - Categorical
##### ridageyr - Age in years at screening - Continuous / Numerical
##### ridreth1 - Race/Hispanic origin - Categorical
##### dmdeduc2 - Education level - Adults 20+ - Categorical
##### dmdmartl - Marital status - Categorical
##### indhhin2 - Annual household income - Categorical
##### bmxbmi - Body Mass Index (kg/m**2) - Continuous / Numerical
##### diq010 - Doctor diagnosed diabetes - Categorical / Target
##### lbxglu - Fasting Glucose (mg/dL) - Continuous / Numerical\(~\)
\(~\)
\(~\)
21.1.1 Install if not Function
install_if_not <- function( list.of.packages ) {
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) { install.packages(new.packages) } else { print(paste0("the package '", list.of.packages , "' is already installed")) }
}\(~\)
\(~\)
\(~\)
\(~\)
21.2 Data Prep
One thing we notice is there are a large number of missing values, take for lbxglu or example. For this example we will omit any values that have an ‘NA’ value, but we could also employ a missing value imputation strategy:
21.2.1 EDA and Imputation
library('tidyverse')── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsdiab_pop %>%
mutate( lbxglu_miss = ifelse(is.na(lbxglu),"missing","reported_value") ) %>%
group_by(lbxglu_miss) %>%
summarise( cnt= n() )# A tibble: 2 × 2
lbxglu_miss cnt
<chr> <int>
1 missing 3267
2 reported_value 2452# We could impute these values with 0 and add a flag indicating so:
diab_pop_impute0glu <- diab_pop %>%
mutate( lbxglu_miss = ifelse(is.na(lbxglu),"imputed_with_0","reported_value") ) %>%
mutate( lbxglu = ifelse(is.na(lbxglu),0,lbxglu) )
glimpse(diab_pop_impute0glu)Rows: 5,719
Columns: 11
$ seqn <dbl> 83732, 83733, 83734, 83735, 83736, 83737, 83741, 83742, 83…
$ riagendr <fct> Male, Male, Male, Female, Female, Female, Male, Female, Ma…
$ ridageyr <dbl> 62, 53, 78, 56, 42, 72, 22, 32, 56, 46, 45, 30, 67, 67, 57…
$ ridreth1 <fct> Non-Hispanic White, Non-Hispanic White, Non-Hispanic White…
$ dmdeduc2 <fct> College grad or above, High school graduate/GED, High scho…
$ dmdmartl <fct> Married, Divorced, Married, Living with partner, Divorced,…
$ indhhin2 <fct> "$65,000-$74,999", "$15,000-$19,999", "$20,000-$24,999", "…
$ bmxbmi <dbl> 27.8, 30.8, 28.8, 42.4, 20.3, 28.6, 28.0, 28.2, 33.6, 27.6…
$ diq010 <fct> Diabetes, No Diabetes, Diabetes, No Diabetes, No Diabetes,…
$ lbxglu <dbl> 0, 101, 84, 0, 84, 107, 95, 0, 0, 0, 84, 0, 130, 284, 398,…
$ lbxglu_miss <chr> "imputed_with_0", "reported_value", "reported_value", "imp…# For this example we will omit any rows with any missing values:
diab_pop.no_na_vals <- diab_pop %>% na.omit()
glimpse(diab_pop.no_na_vals)Rows: 1,876
Columns: 10
$ seqn <dbl> 83733, 83734, 83737, 83750, 83754, 83755, 83757, 83761, 83787…
$ riagendr <fct> Male, Male, Female, Male, Female, Male, Female, Female, Femal…
$ ridageyr <dbl> 53, 78, 72, 45, 67, 67, 57, 24, 68, 66, 56, 37, 20, 24, 80, 7…
$ ridreth1 <fct> Non-Hispanic White, Non-Hispanic White, MexicanAmerican, Othe…
$ dmdeduc2 <fct> High school graduate/GED, High school graduate/GED, Grades 9-…
$ dmdmartl <fct> Divorced, Married, Separated, Never married, Married, Widowed…
$ indhhin2 <fct> "$15,000-$19,999", "$20,000-$24,999", "$75,000-$99,999", "$65…
$ bmxbmi <dbl> 30.8, 28.8, 28.6, 24.1, 43.7, 28.8, 35.4, 25.3, 33.5, 34.0, 2…
$ diq010 <fct> No Diabetes, Diabetes, No Diabetes, No Diabetes, No Diabetes,…
$ lbxglu <dbl> 101, 84, 107, 84, 130, 284, 398, 95, 111, 113, 397, 100, 94, …\(~\)
\(~\)
\(~\)
21.3 Split Data with caret
We will want to split our data into two main sets: a training set to train the model and a testing set used to estimate model performance metrics.
install_if_not('caret')[1] "the package 'caret' is already installed"library('caret')Loading required package: lattice
Attaching package: 'caret'The following object is masked from 'package:purrr':
lift# this will ensure our results are the same every run, to randomize you may use: `set.seed(Sys.time())` or `set.seed(runif(1))`
set.seed(8675309)
# The createDataPartition function is used to create training and test sets
trainIndex <- createDataPartition(diab_pop.no_na_vals$diq010,
p = .6,
list = FALSE,
times = 1)\(~\)
\(~\)
21.3.1 Define the Training Set
diab_pop.no_na_vals.train <- diab_pop.no_na_vals[trainIndex, ]
# Notice the size of the overall dataset
dim(diab_pop.no_na_vals)[1] 1876 10# and the size of our training set:
.6*nrow(diab_pop.no_na_vals) [1] 1125.6nrow(diab_pop.no_na_vals.train)[1] 1126\(~\)
\(~\)
21.3.2 Define the Testing Set
diab_pop.no_na_vals.test <- diab_pop.no_na_vals[-trainIndex, ]
nrow(diab_pop.no_na_vals) - .6*nrow(diab_pop.no_na_vals) [1] 750.4dim(diab_pop.no_na_vals.test)[1] 750 10\(~\)
\(~\)
\(~\)
\(~\)
21.4 Fit Decision Trees with rpart
train_set <- diab_pop.no_na_vals.train
install_if_not('rpart')[1] "the package 'rpart' is already installed"library('rpart')
### diq010 ~ riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu
### diq010 ~ ridreth1 + lbxglu
tree_1 <- rpart(diq010 ~ riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu,
data = train_set,
method="class",
#parms = list(split = 'information'),
control = rpart.control(minsplit = 1,
minbucket = 1, #round(minsplit/3)
cp = 0.006, #3*10^(-3),
maxcompete = 4,
maxsurrogate = 5,
usesurrogate = 2,
xval = 10,
surrogatestyle = 0,
maxdepth = 30))
tree_1n= 1126
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 1126 169 No Diabetes (0.15008881 0.84991119)
2) lbxglu>=135 132 31 Diabetes (0.76515152 0.23484848)
4) lbxglu>=154.5 96 13 Diabetes (0.86458333 0.13541667)
8) indhhin2=$0-$4,999,$5,000-$9,999,$10,000-$14,999,$20,000-$24,999,$25,000-$34,999,$45,000-$54,999,$65,000-$74,999,$75,000-$99,999,$100,000+ 78 7 Diabetes (0.91025641 0.08974359) *
9) indhhin2=$15,000-$19,999,20,000+,less than $20,000 18 6 Diabetes (0.66666667 0.33333333)
18) ridageyr>=49 15 3 Diabetes (0.80000000 0.20000000)
36) bmxbmi< 39.4 13 1 Diabetes (0.92307692 0.07692308) *
37) bmxbmi>=39.4 2 0 No Diabetes (0.00000000 1.00000000) *
19) ridageyr< 49 3 0 No Diabetes (0.00000000 1.00000000) *
5) lbxglu< 154.5 36 18 Diabetes (0.50000000 0.50000000)
10) indhhin2=$25,000-$34,999,$65,000-$74,999,20,000+,$100,000+ 13 2 Diabetes (0.84615385 0.15384615) *
11) indhhin2=$5,000-$9,999,$10,000-$14,999,$15,000-$19,999,$20,000-$24,999,$45,000-$54,999,less than $20,000,$75,000-$99,999 23 7 No Diabetes (0.30434783 0.69565217)
22) dmdmartl=Married,Divorced 14 7 Diabetes (0.50000000 0.50000000)
44) ridageyr>=63.5 8 2 Diabetes (0.75000000 0.25000000) *
45) ridageyr< 63.5 6 1 No Diabetes (0.16666667 0.83333333) *
23) dmdmartl=Widowed,Never married,Living with partner 9 0 No Diabetes (0.00000000 1.00000000) *
3) lbxglu< 135 994 68 No Diabetes (0.06841046 0.93158954)
6) lbxglu>=113.5 146 34 No Diabetes (0.23287671 0.76712329)
12) indhhin2=$0-$4,999,$5,000-$9,999,$20,000-$24,999,less than $20,000 35 17 No Diabetes (0.48571429 0.51428571)
24) bmxbmi>=30.55 20 7 Diabetes (0.65000000 0.35000000)
48) lbxglu< 132 18 5 Diabetes (0.72222222 0.27777778) *
49) lbxglu>=132 2 0 No Diabetes (0.00000000 1.00000000) *
25) bmxbmi< 30.55 15 4 No Diabetes (0.26666667 0.73333333)
50) dmdmartl=Married,Separated 6 2 Diabetes (0.66666667 0.33333333)
100) bmxbmi< 26.8 4 0 Diabetes (1.00000000 0.00000000) *
101) bmxbmi>=26.8 2 0 No Diabetes (0.00000000 1.00000000) *
51) dmdmartl=Widowed,Divorced,Never married,Living with partner 9 0 No Diabetes (0.00000000 1.00000000) *
13) indhhin2=$10,000-$14,999,$15,000-$19,999,$25,000-$34,999,$45,000-$54,999,$65,000-$74,999,20,000+,$75,000-$99,999,$100,000+ 111 17 No Diabetes (0.15315315 0.84684685)
26) ridageyr>=73.5 18 7 No Diabetes (0.38888889 0.61111111)
52) lbxglu>=124 6 1 Diabetes (0.83333333 0.16666667) *
53) lbxglu< 124 12 2 No Diabetes (0.16666667 0.83333333) *
27) ridageyr< 73.5 93 10 No Diabetes (0.10752688 0.89247312) *
7) lbxglu< 113.5 848 34 No Diabetes (0.04009434 0.95990566)
14) lbxglu< 78.5 19 6 No Diabetes (0.31578947 0.68421053)
28) indhhin2=$0-$4,999,$5,000-$9,999 3 0 Diabetes (1.00000000 0.00000000) *
29) indhhin2=$15,000-$19,999,$20,000-$24,999,$25,000-$34,999,$45,000-$54,999,$65,000-$74,999,20,000+,less than $20,000,$75,000-$99,999,$100,000+ 16 3 No Diabetes (0.18750000 0.81250000)
58) ridageyr>=56 6 3 Diabetes (0.50000000 0.50000000)
116) ridageyr< 62.5 3 0 Diabetes (1.00000000 0.00000000) *
117) ridageyr>=62.5 3 0 No Diabetes (0.00000000 1.00000000) *
59) ridageyr< 56 10 0 No Diabetes (0.00000000 1.00000000) *
15) lbxglu>=78.5 829 28 No Diabetes (0.03377563 0.96622437) *plot(tree_1)
\(~\)
\(~\)
21.5 Better View with rpart.plot
install_if_not('rpart.plot')[1] "the package 'rpart.plot' is already installed"library('rpart.plot')
rpart.plot(tree_1)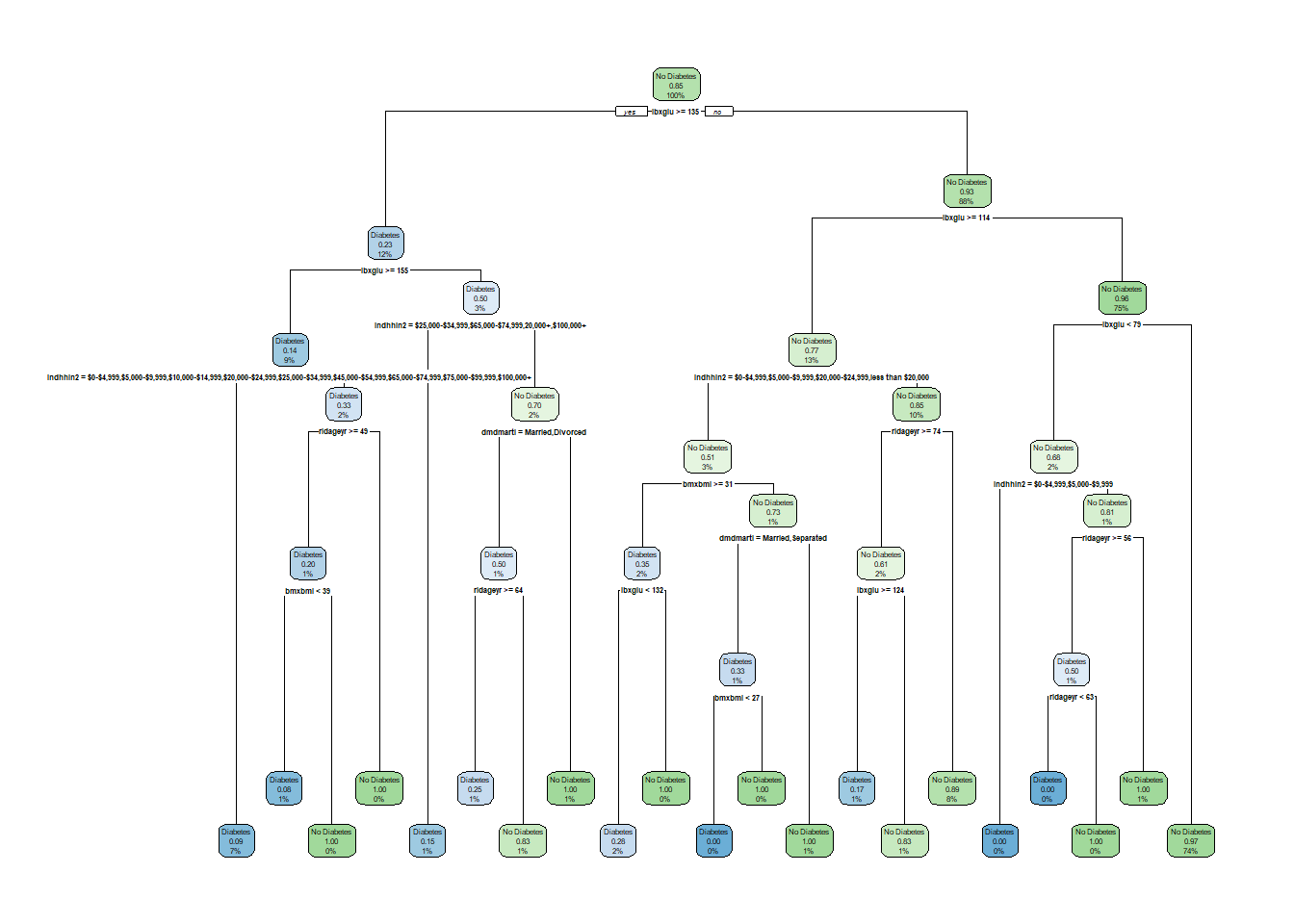
\(~\)
\(~\)
21.6 rpart output
str(tree_1,1)List of 15
$ frame :'data.frame': 41 obs. of 9 variables:
$ where : Named int [1:1126] 41 41 32 4 41 7 41 41 41 31 ...
..- attr(*, "names")= chr [1:1126] "2" "11" "13" "14" ...
$ call : language rpart(formula = diq010 ~ riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu, | __truncated__ ...
$ terms :Classes 'terms', 'formula' language diq010 ~ riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu
.. ..- attr(*, "variables")= language list(diq010, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "factors")= int [1:9, 1:8] 0 1 0 0 0 0 0 0 0 0 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. ..- attr(*, "term.labels")= chr [1:8] "riagendr" "ridageyr" "ridreth1" "dmdeduc2" ...
.. ..- attr(*, "order")= int [1:8] 1 1 1 1 1 1 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(diq010, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "dataClasses")= Named chr [1:9] "factor" "factor" "numeric" "factor" ...
.. .. ..- attr(*, "names")= chr [1:9] "diq010" "riagendr" "ridageyr" "ridreth1" ...
$ cptable : num [1:5, 1:5] 0.4142 0.02663 0.01183 0.00888 0.006 ...
..- attr(*, "dimnames")=List of 2
$ method : chr "class"
$ parms :List of 3
$ control :List of 9
$ functions :List of 3
$ numresp : int 4
$ splits : num [1:144, 1:5] 1126 1126 1126 1126 1126 ...
..- attr(*, "dimnames")=List of 2
$ csplit : int [1:78, 1:14] 1 1 1 1 1 1 3 3 1 3 ...
$ variable.importance: Named num [1:8] 142.17 19.7 14.66 14.46 8.14 ...
..- attr(*, "names")= chr [1:8] "lbxglu" "indhhin2" "ridageyr" "bmxbmi" ...
$ y : int [1:1126] 2 2 2 1 2 2 2 2 2 2 ...
$ ordered : Named logi [1:8] FALSE FALSE FALSE FALSE FALSE FALSE ...
..- attr(*, "names")= chr [1:8] "riagendr" "ridageyr" "ridreth1" "dmdeduc2" ...
- attr(*, "xlevels")=List of 5
- attr(*, "ylevels")= chr [1:2] "Diabetes" "No Diabetes"
- attr(*, "class")= chr "rpart"tree_1$splits count ncat improve index adj
lbxglu 1126 1 113.1344112 135.00 0.00000000
ridageyr 1126 1 22.8255083 48.50 0.00000000
bmxbmi 1126 1 7.6602898 27.65 0.00000000
dmdeduc2 1126 5 5.7790434 1.00 0.00000000
dmdmartl 1126 6 5.7449142 2.00 0.00000000
lbxglu 132 1 6.9602273 154.50 0.00000000
bmxbmi 132 1 4.1704107 25.25 0.00000000
indhhin2 132 14 2.8812328 3.00 0.00000000
ridageyr 132 1 2.3778555 27.50 0.00000000
dmdmartl 132 6 1.8782314 4.00 0.00000000
ridageyr 0 1 0.7424242 27.50 0.05555556
bmxbmi 0 1 0.7424242 20.85 0.05555556
indhhin2 0 14 0.7348485 5.00 0.02777778
indhhin2 96 14 1.7355769 6.00 0.00000000
bmxbmi 96 -1 1.0405963 37.20 0.00000000
ridreth1 96 5 0.5720238 7.00 0.00000000
dmdmartl 96 6 0.5628608 8.00 0.00000000
ridageyr 96 1 0.5208333 61.50 0.00000000
bmxbmi 0 1 0.8333333 21.35 0.11111111
lbxglu 0 -1 0.8333333 393.50 0.11111111
ridageyr 18 1 3.2000000 49.00 0.00000000
bmxbmi 18 -1 3.2000000 39.40 0.00000000
dmdmartl 18 6 2.0000000 9.00 0.00000000
lbxglu 18 1 0.8000000 419.50 0.00000000
ridreth1 18 5 0.5000000 10.00 0.00000000
bmxbmi 15 -1 2.9538462 39.40 0.00000000
ridageyr 15 1 1.3714286 62.00 0.00000000
dmdmartl 15 6 1.3714286 11.00 0.00000000
dmdeduc2 15 5 0.6000000 12.00 0.00000000
ridreth1 15 5 0.4363636 13.00 0.00000000
indhhin2 36 14 4.8762542 14.00 0.00000000
dmdmartl 36 6 4.4307692 15.00 0.00000000
ridageyr 36 1 2.8928571 48.00 0.00000000
bmxbmi 36 1 2.8928571 25.35 0.00000000
dmdeduc2 36 5 1.0451613 16.00 0.00000000
dmdeduc2 0 5 0.6944444 17.00 0.15384615
bmxbmi 0 1 0.6944444 40.40 0.15384615
lbxglu 0 1 0.6666667 146.50 0.07692308
dmdmartl 23 6 2.7391304 18.00 0.00000000
ridageyr 23 1 1.9209486 63.50 0.00000000
ridreth1 23 5 1.8641304 19.00 0.00000000
dmdeduc2 23 5 0.8970252 20.00 0.00000000
bmxbmi 23 -1 0.8970252 37.60 0.00000000
ridreth1 0 5 0.7391304 21.00 0.33333333
ridageyr 0 1 0.6956522 32.00 0.22222222
dmdeduc2 0 5 0.6956522 22.00 0.22222222
indhhin2 0 14 0.6956522 23.00 0.22222222
bmxbmi 0 -1 0.6956522 31.05 0.22222222
ridageyr 14 1 2.3333333 63.50 0.00000000
lbxglu 14 -1 1.4000000 142.50 0.00000000
ridreth1 14 5 1.1666667 24.00 0.00000000
dmdmartl 14 6 1.1666667 25.00 0.00000000
bmxbmi 14 -1 1.1666667 37.80 0.00000000
ridreth1 0 5 0.7142857 26.00 0.33333333
dmdeduc2 0 5 0.7142857 27.00 0.33333333
indhhin2 0 14 0.7142857 28.00 0.33333333
bmxbmi 0 -1 0.7142857 37.80 0.33333333
riagendr 0 2 0.6428571 29.00 0.16666667
lbxglu 994 1 9.2582086 113.50 0.00000000
ridageyr 994 1 4.8704224 48.50 0.00000000
indhhin2 994 14 3.7854728 30.00 0.00000000
dmdeduc2 994 5 2.3395179 31.00 0.00000000
bmxbmi 994 1 2.1377841 48.55 0.00000000
indhhin2 146 14 5.8858765 32.00 0.00000000
ridageyr 146 1 2.3689223 73.50 0.00000000
dmdeduc2 146 5 1.9580029 33.00 0.00000000
lbxglu 146 1 1.5740855 121.50 0.00000000
ridreth1 146 5 1.5468950 34.00 0.00000000
bmxbmi 0 -1 0.7739726 22.25 0.05714286
bmxbmi 35 1 2.5190476 30.55 0.00000000
ridreth1 35 5 1.7654346 35.00 0.00000000
dmdmartl 35 6 1.7210084 36.00 0.00000000
dmdeduc2 35 5 1.6822955 37.00 0.00000000
ridageyr 35 1 1.2190476 59.50 0.00000000
dmdeduc2 0 5 0.7428571 38.00 0.40000000
ridreth1 0 5 0.7142857 39.00 0.33333333
dmdmartl 0 6 0.6857143 40.00 0.26666667
lbxglu 0 1 0.6571429 121.00 0.20000000
ridageyr 0 -1 0.6285714 74.50 0.13333333
lbxglu 20 -1 1.8777778 132.00 0.00000000
ridreth1 20 5 1.2250000 41.00 0.00000000
dmdeduc2 20 5 0.8894737 42.00 0.00000000
indhhin2 20 14 0.8894737 43.00 0.00000000
bmxbmi 20 1 0.8647059 43.40 0.00000000
dmdmartl 15 6 3.2000000 44.00 0.00000000
ridageyr 15 1 1.4222222 61.50 0.00000000
dmdeduc2 15 5 1.4222222 45.00 0.00000000
lbxglu 15 -1 1.1523810 114.50 0.00000000
bmxbmi 15 -1 1.0666667 26.80 0.00000000
indhhin2 0 14 0.7333333 46.00 0.33333333
riagendr 0 2 0.6666667 47.00 0.16666667
ridageyr 0 1 0.6666667 61.50 0.16666667
dmdeduc2 0 5 0.6666667 48.00 0.16666667
bmxbmi 0 1 0.6666667 22.70 0.16666667
bmxbmi 6 -1 2.6666667 26.80 0.00000000
ridageyr 6 1 1.0666667 59.00 0.00000000
ridreth1 6 5 1.0666667 49.00 0.00000000
dmdeduc2 6 5 1.0666667 50.00 0.00000000
riagendr 6 2 0.2666667 51.00 0.00000000
ridageyr 111 1 2.3877749 73.50 0.00000000
lbxglu 111 1 1.4795233 121.50 0.00000000
dmdeduc2 111 5 1.2573089 52.00 0.00000000
dmdmartl 111 6 0.8262812 53.00 0.00000000
indhhin2 111 14 0.8245388 54.00 0.00000000
dmdmartl 0 6 0.8738739 55.00 0.22222222
lbxglu 18 1 3.5555556 124.00 0.00000000
dmdeduc2 18 5 2.7777778 56.00 0.00000000
dmdmartl 18 6 2.7777778 57.00 0.00000000
riagendr 18 2 1.3867244 58.00 0.00000000
ridreth1 18 5 1.3412698 59.00 0.00000000
ridreth1 0 5 0.7222222 60.00 0.16666667
indhhin2 0 14 0.7222222 61.00 0.16666667
lbxglu 848 -1 2.9544941 78.50 0.00000000
ridageyr 848 1 1.9956934 55.50 0.00000000
bmxbmi 848 1 1.5644073 48.85 0.00000000
indhhin2 848 14 1.1369908 62.00 0.00000000
dmdmartl 848 6 1.0245136 63.00 0.00000000
indhhin2 19 14 3.3355263 64.00 0.00000000
bmxbmi 19 1 3.1819549 29.45 0.00000000
dmdmartl 19 6 2.0928793 65.00 0.00000000
ridageyr 19 1 1.9660819 47.50 0.00000000
dmdeduc2 19 5 1.9105263 66.00 0.00000000
bmxbmi 0 1 0.8947368 29.45 0.33333333
lbxglu 0 1 0.8947368 77.50 0.33333333
ridageyr 16 1 1.8750000 56.00 0.00000000
dmdeduc2 16 5 1.6955128 67.00 0.00000000
bmxbmi 16 -1 1.4083333 20.30 0.00000000
lbxglu 16 -1 1.4083333 57.00 0.00000000
ridreth1 16 5 1.1250000 68.00 0.00000000
lbxglu 0 -1 0.8750000 72.50 0.66666667
dmdeduc2 0 5 0.8125000 69.00 0.50000000
indhhin2 0 14 0.7500000 70.00 0.33333333
bmxbmi 0 -1 0.7500000 21.80 0.33333333
ridreth1 0 5 0.6875000 71.00 0.16666667
ridageyr 6 -1 3.0000000 62.50 0.00000000
indhhin2 6 14 3.0000000 72.00 0.00000000
ridreth1 6 5 1.5000000 73.00 0.00000000
dmdeduc2 6 5 0.6000000 74.00 0.00000000
dmdmartl 6 6 0.6000000 75.00 0.00000000
riagendr 0 2 0.6666667 76.00 0.33333333
dmdeduc2 0 5 0.6666667 77.00 0.33333333
dmdmartl 0 6 0.6666667 78.00 0.33333333
bmxbmi 0 1 0.6666667 24.65 0.33333333
lbxglu 0 1 0.6666667 68.50 0.33333333tree_1$cptable CP nsplit rel error xerror xstd
1 0.41420118 0 1.0000000 1.0000000 0.07091587
2 0.02662722 1 0.5857988 0.6272189 0.05798252
3 0.01183432 3 0.5325444 0.6331361 0.05822682
4 0.00887574 13 0.4142012 0.6982249 0.06081565
5 0.00600000 20 0.3491124 0.7455621 0.06259352\(~\)
\(~\)
\(~\)
21.7 Prune Decision Tree
library('tidyverse')
tree_1_cptable_tb <- as_tibble(tree_1$cptable)
tree_1_cptable_tb# A tibble: 5 × 5
CP nsplit `rel error` xerror xstd
<dbl> <dbl> <dbl> <dbl> <dbl>
1 0.414 0 1 1 0.0709
2 0.0266 1 0.586 0.627 0.0580
3 0.0118 3 0.533 0.633 0.0582
4 0.00888 13 0.414 0.698 0.0608
5 0.006 20 0.349 0.746 0.0626cp_val <- (tree_1_cptable_tb %>%
arrange(-CP) %>%
filter(row_number()==2))$CP
cp_val[1] 0.02662722tree_prune <- prune(tree_1, cp = cp_val)
tree_prunen= 1126
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 1126 169 No Diabetes (0.15008881 0.84991119)
2) lbxglu>=135 132 31 Diabetes (0.76515152 0.23484848) *
3) lbxglu< 135 994 68 No Diabetes (0.06841046 0.93158954) *rpart.plot(tree_prune)$cptable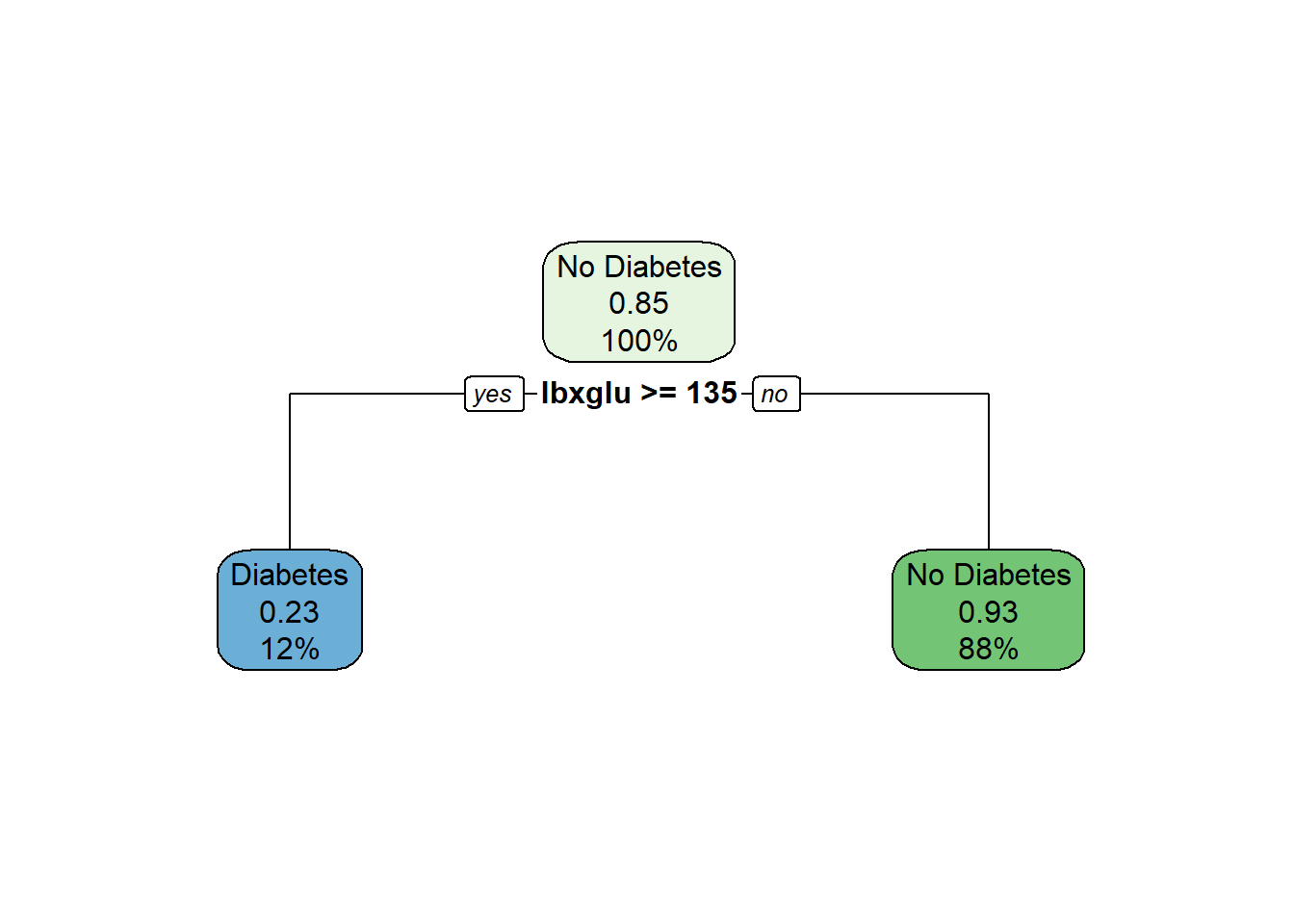
NULLtree_prune$cptable CP nsplit rel error xerror xstd
1 0.41420118 0 1.0000000 1.0000000 0.07091587
2 0.02662722 1 0.5857988 0.6272189 0.05798252str(tree_prune)List of 15
$ frame :'data.frame': 3 obs. of 9 variables:
..$ var : chr [1:3] "lbxglu" "<leaf>" "<leaf>"
..$ n : int [1:3] 1126 132 994
..$ wt : num [1:3] 1126 132 994
..$ dev : num [1:3] 169 31 68
..$ yval : num [1:3] 2 1 2
..$ complexity: num [1:3] 0.4142 0.0266 0.0118
..$ ncompete : int [1:3] 4 0 0
..$ nsurrogate: int [1:3] 0 0 0
..$ yval2 : num [1:3, 1:6] 2 1 2 169 101 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : NULL
.. .. ..$ : chr [1:6] "" "" "" "" ...
$ where : int [1:1126] 3 3 3 2 3 2 3 3 3 3 ...
$ call : language rpart(formula = diq010 ~ riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu, | __truncated__ ...
$ terms :Classes 'terms', 'formula' language diq010 ~ riagendr + ridageyr + ridreth1 + dmdeduc2 + dmdmartl + indhhin2 + bmxbmi + lbxglu
.. ..- attr(*, "variables")= language list(diq010, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "factors")= int [1:9, 1:8] 0 1 0 0 0 0 0 0 0 0 ...
.. .. ..- attr(*, "dimnames")=List of 2
.. .. .. ..$ : chr [1:9] "diq010" "riagendr" "ridageyr" "ridreth1" ...
.. .. .. ..$ : chr [1:8] "riagendr" "ridageyr" "ridreth1" "dmdeduc2" ...
.. ..- attr(*, "term.labels")= chr [1:8] "riagendr" "ridageyr" "ridreth1" "dmdeduc2" ...
.. ..- attr(*, "order")= int [1:8] 1 1 1 1 1 1 1 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(diq010, riagendr, ridageyr, ridreth1, dmdeduc2, dmdmartl, indhhin2, bmxbmi, lbxglu)
.. ..- attr(*, "dataClasses")= Named chr [1:9] "factor" "factor" "numeric" "factor" ...
.. .. ..- attr(*, "names")= chr [1:9] "diq010" "riagendr" "ridageyr" "ridreth1" ...
$ cptable : num [1:2, 1:5] 0.4142 0.0266 0 1 1 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:2] "1" "2"
.. ..$ : chr [1:5] "CP" "nsplit" "rel error" "xerror" ...
$ method : chr "class"
$ parms :List of 3
..$ prior: num [1:2(1d)] 0.15 0.85
.. ..- attr(*, "dimnames")=List of 1
.. .. ..$ : chr [1:2] "1" "2"
..$ loss : num [1:2, 1:2] 0 1 1 0
..$ split: num 1
$ control :List of 9
..$ minsplit : num 1
..$ minbucket : num 1
..$ cp : num 0.006
..$ maxcompete : num 4
..$ maxsurrogate : num 5
..$ usesurrogate : num 2
..$ surrogatestyle: num 0
..$ maxdepth : num 30
..$ xval : num 10
$ functions :List of 3
..$ summary:function (yval, dev, wt, ylevel, digits)
..$ print :function (yval, ylevel, digits, nsmall)
..$ text :function (yval, dev, wt, ylevel, digits, n, use.n)
$ numresp : int 4
$ splits : num [1:5, 1:5] 1126 1126 1126 1126 1126 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:5] "lbxglu" "ridageyr" "bmxbmi" "dmdeduc2" ...
.. ..$ : chr [1:5] "count" "ncat" "improve" "index" ...
$ csplit : int [1:2, 1:14] 1 1 3 1 3 1 3 1 3 3 ...
$ variable.importance: Named num 113
..- attr(*, "names")= chr "lbxglu"
$ y : int [1:1126] 2 2 2 1 2 2 2 2 2 2 ...
$ ordered : Named logi [1:8] FALSE FALSE FALSE FALSE FALSE FALSE ...
..- attr(*, "names")= chr [1:8] "riagendr" "ridageyr" "ridreth1" "dmdeduc2" ...
- attr(*, "xlevels")=List of 5
..$ riagendr: chr [1:2] "Male" "Female"
..$ ridreth1: chr [1:5] "MexicanAmerican" "Other Hispanic" "Non-Hispanic White" "Non-Hispanic Black" ...
..$ dmdeduc2: chr [1:5] "Less than 9th grade" "Grades 9-11th" "High school graduate/GED" "Some college or AA degrees" ...
..$ dmdmartl: chr [1:6] "Married" "Widowed" "Divorced" "Separated" ...
..$ indhhin2: chr [1:14] "$0-$4,999" "$5,000-$9,999" "$10,000-$14,999" "$15,000-$19,999" ...
- attr(*, "ylevels")= chr [1:2] "Diabetes" "No Diabetes"
- attr(*, "class")= chr "rpart"lbxglu_split_level <- tree_prune$splits['lbxglu','index']\(~\)
\(~\)
\(~\)
21.8 Score Decision Tree Model on Training Set
\(~\)
\(~\)
21.8.1 Score Output Probabilities
y_hat_probs <- predict(tree_prune, train_set)
head(y_hat_probs) Diabetes No Diabetes
2 0.06841046 0.9315895
11 0.06841046 0.9315895
13 0.06841046 0.9315895
14 0.76515152 0.2348485
29 0.06841046 0.9315895
32 0.76515152 0.2348485str(y_hat_probs) num [1:1126, 1:2] 0.0684 0.0684 0.0684 0.7652 0.0684 ...
- attr(*, "dimnames")=List of 2
..$ : chr [1:1126] "2" "11" "13" "14" ...
..$ : chr [1:2] "Diabetes" "No Diabetes"\(~\)
\(~\)
21.8.2 Score Output Class
y_hat_class <- predict(tree_prune, train_set, type ="class")
head(y_hat_class) 2 11 13 14 29 32
No Diabetes No Diabetes No Diabetes Diabetes No Diabetes Diabetes
Levels: Diabetes No Diabetesstr(y_hat_class) Factor w/ 2 levels "Diabetes","No Diabetes": 2 2 2 1 2 1 2 2 2 2 ...
- attr(*, "names")= chr [1:1126] "2" "11" "13" "14" ...\(~\)
\(~\)
21.8.3 View Training Dataset with Scores
train.scored <- as_tibble(cbind(train_set, y_hat_probs, y_hat_class))
glimpse(train.scored)Rows: 1,126
Columns: 13
$ seqn <dbl> 83733, 83750, 83754, 83755, 83787, 83790, 83799, 83809, …
$ riagendr <fct> Male, Male, Female, Male, Female, Male, Female, Female, …
$ ridageyr <dbl> 53, 45, 67, 67, 68, 56, 37, 20, 24, 80, 39, 35, 40, 74, …
$ ridreth1 <fct> Non-Hispanic White, Other, Other Hispanic, Non-Hispanic …
$ dmdeduc2 <fct> High school graduate/GED, Grades 9-11th, College grad or…
$ dmdmartl <fct> Divorced, Never married, Married, Widowed, Divorced, Mar…
$ indhhin2 <fct> "$15,000-$19,999", "$65,000-$74,999", "$25,000-$34,999",…
$ bmxbmi <dbl> 30.8, 24.1, 43.7, 28.8, 33.5, 24.4, 25.5, 26.2, 26.9, 28…
$ diq010 <fct> No Diabetes, No Diabetes, No Diabetes, Diabetes, No Diab…
$ lbxglu <dbl> 101, 84, 130, 284, 111, 397, 100, 94, 105, 119, 101, 97,…
$ Diabetes <dbl> 0.06841046, 0.06841046, 0.06841046, 0.76515152, 0.068410…
$ `No Diabetes` <dbl> 0.9315895, 0.9315895, 0.9315895, 0.2348485, 0.9315895, 0…
$ y_hat_class <fct> No Diabetes, No Diabetes, No Diabetes, Diabetes, No Diab…\(~\)
\(~\)
\(~\)
\(~\)
21.9 Why was 135 chosen as the split value of lbxglu
The goal in some of these subsequent sections is to give some insight as to how the decision tree chooses to make the cut.
\(~\)
\(~\)
21.9.1 Compare Confusion Matrix and 2-by-2 Tables
library('caret')
cm_1 <- confusionMatrix( data = train.scored$y_hat_class,
reference = train.scored$diq010,
positive = 'Diabetes',
mode = "everything")
cm_1Confusion Matrix and Statistics
Reference
Prediction Diabetes No Diabetes
Diabetes 101 31
No Diabetes 68 926
Accuracy : 0.9121
95% CI : (0.894, 0.928)
No Information Rate : 0.8499
P-Value [Acc > NIR] : 2.918e-10
Kappa : 0.6212
Mcnemar's Test P-Value : 0.0002967
Sensitivity : 0.5976
Specificity : 0.9676
Pos Pred Value : 0.7652
Neg Pred Value : 0.9316
Precision : 0.7652
Recall : 0.5976
F1 : 0.6711
Prevalence : 0.1501
Detection Rate : 0.0897
Detection Prevalence : 0.1172
Balanced Accuracy : 0.7826
'Positive' Class : Diabetes
str(cm_1)List of 6
$ positive: chr "Diabetes"
$ table : 'table' int [1:2, 1:2] 101 68 31 926
..- attr(*, "dimnames")=List of 2
.. ..$ Prediction: chr [1:2] "Diabetes" "No Diabetes"
.. ..$ Reference : chr [1:2] "Diabetes" "No Diabetes"
$ overall : Named num [1:7] 0.912 0.621 0.894 0.928 0.85 ...
..- attr(*, "names")= chr [1:7] "Accuracy" "Kappa" "AccuracyLower" "AccuracyUpper" ...
$ byClass : Named num [1:11] 0.598 0.968 0.765 0.932 0.765 ...
..- attr(*, "names")= chr [1:11] "Sensitivity" "Specificity" "Pos Pred Value" "Neg Pred Value" ...
$ mode : chr "everything"
$ dots : list()
- attr(*, "class")= chr "confusionMatrix"cm_1$table Reference
Prediction Diabetes No Diabetes
Diabetes 101 31
No Diabetes 68 926cm_1$byClass Sensitivity Specificity Pos Pred Value
0.59763314 0.96760711 0.76515152
Neg Pred Value Precision Recall
0.93158954 0.76515152 0.59763314
F1 Prevalence Detection Rate
0.67109635 0.15008881 0.08969805
Detection Prevalence Balanced Accuracy
0.11722913 0.78262012 table_1 <- table(train.scored$y_hat_class, train.scored$diq010)
table_1
Diabetes No Diabetes
Diabetes 101 31
No Diabetes 68 926cm_1$table Reference
Prediction Diabetes No Diabetes
Diabetes 101 31
No Diabetes 68 926gplots::balloonplot(cm_1$table,
main ="Balloon Plot for lbxglu_flag by Diabetes \n Area is proportional to Freq.")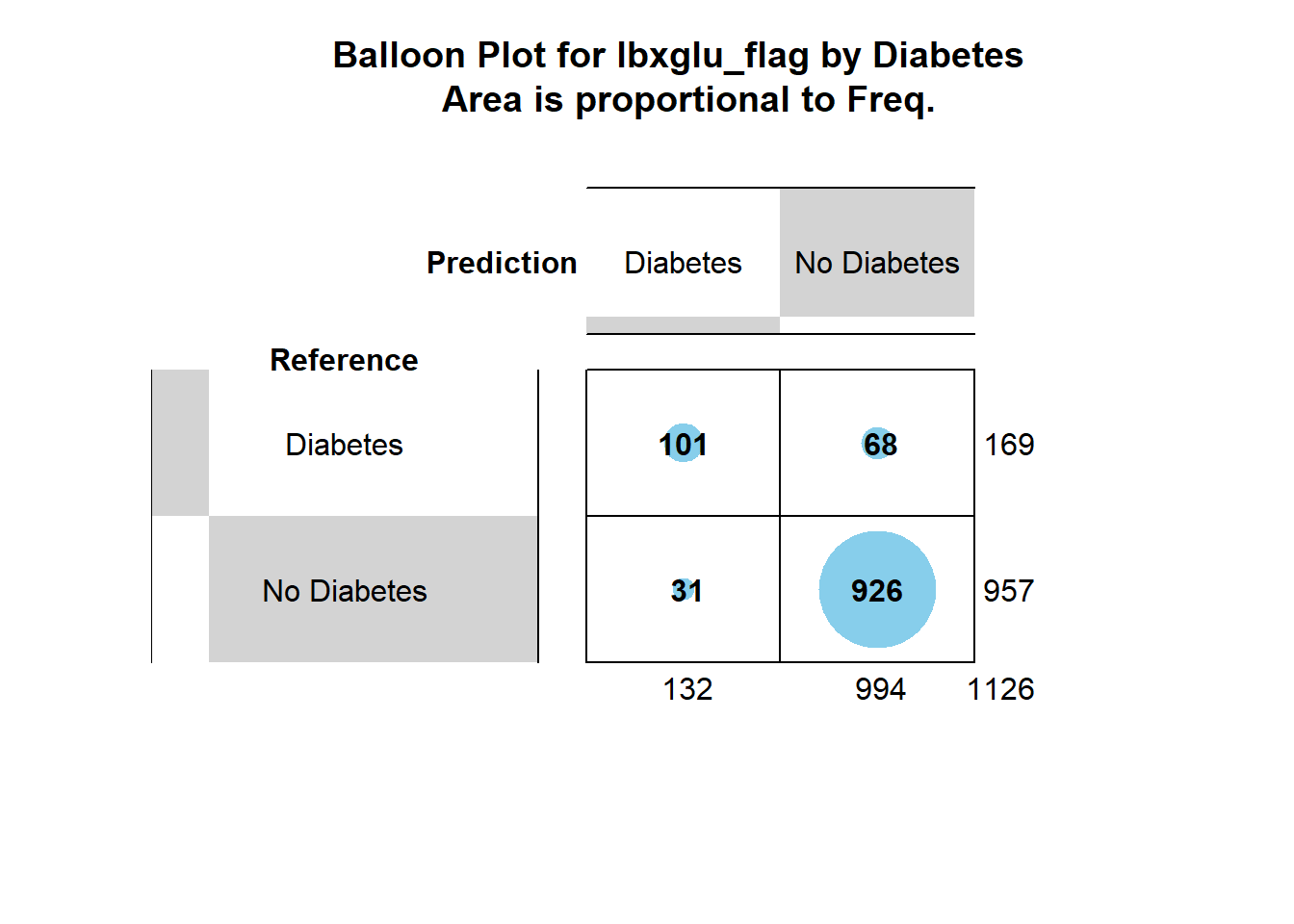
chisq.test(cm_1$table)$p.value[1] 2.944895e-97\(~\)
\(~\)
\(~\)
21.9.2 Programming a Confusion Matrix from a 2-by-2 Table
table_1 <- table(train.scored$y_hat_class, train.scored$diq010)
table_1
Diabetes No Diabetes
Diabetes 101 31
No Diabetes 68 926TP <- table_1[1,1]
FP <- table_1[1,2]
FN <- table_1[2,1]
TN <- table_1[2,2]
TPR = TP / (TP+FN)
TNR = TN / (TN+FP)
PPV = TP / (TP+FP)
NPV = TN / (TN+FN)
ACC = (TP+TN)/(TP+TN+FP+FN)
F1 = 2/((1/TPR) + (1/PPV))\(~\)
\(~\)
\(~\)
21.9.2.1 Check our work
cm_1$byClass['Sensitivity'] - TPRSensitivity
0 cm_1$byClass['Specificity'] - TNRSpecificity
0 cm_1$byClass['Pos Pred Value'] - PPVPos Pred Value
-1.110223e-16 cm_1$overall['Accuracy'] - ACCAccuracy
0 cm_1$byClass['F1'] - F1F1
0 \(~\)
\(~\)
\(~\)
21.10 Decision Tree - Choosing the Cut Point
The goal in some of these subsequent sections is to give some insight as to how the decision tree chooses to make the cut.
This function, will:
- Take in a value for
lbxglu - If the recorded value for
lbxgluis greater than or equal to the input value, then the record is flagged withlbxglu_over_value, otherwise it is flagged withlbxglu_under_value - A 2-by-2 table is then created to mirror the confusion matrix of that decision
- Metrics are reported and returned for that decision:
lbxglu_value_chisq <- function(my_value, return_table=0){
require('tidyverse')
dt <- train_set %>%
mutate(lbxglu_flag = ifelse(lbxglu >= my_value,"lbxglu_over_value","lbxglu_under_value") )
table_1 <- table(dt$lbxglu_flag , dt$diq010)
if(return_table ==1 ){return(table_1)}
TP <- table_1[1,1]
FP <- table_1[1,2]
FN <- table_1[2,1]
TN <- table_1[2,2]
TPR = TP / (TP+FN)
TNR = TN / (TN+FP)
PPV = TP / (TP+FP)
NPV = TN / (TN+FN)
ACC = (TP+TN)/(TP+TN+FP+FN)
F1 = 2/((1/TPR) + (1/PPV))
# GINI AND INFORMATION
base_prob <-table(dt$lbxglu_flag)/length(dt$lbxglu_flag)
crosstab <- table(dt$diq010, dt$lbxglu_flag)
crossprob <- prop.table(crosstab,2)
# GINI
No_Node_Gini <- 1-sum(crossprob[,1]**2)
Yes_Node_Gini <- 1-sum(crossprob[,2]**2)
GINI <- sum(base_prob * c(No_Node_Gini,Yes_Node_Gini))
# INFORMATION
No_Col <- crossprob[crossprob[,1]>0,1]
Yes_Col <- crossprob[crossprob[,2]>0,2]
No_Node_Info <- -sum(No_Col*log(No_Col,2))
Yes_Node_Info <- -sum(Yes_Col*log(Yes_Col,2))
Information <- sum(base_prob * c(No_Node_Info,Yes_Node_Info))
table_1_chisq_pvalue <- tibble::enframe(chisq.test(table_1)$p.value) %>%
rename(chisq_p_value = value) %>%
select(-name) %>%
mutate(lbxglu_value = my_value) %>%
select(lbxglu_value, chisq_p_value) %>%
mutate( TP = TP ) %>%
mutate( FP = FP ) %>%
mutate( FN = FN ) %>%
mutate( TN = TN ) %>%
mutate( PPV = PPV ) %>%
mutate( TPR = TPR ) %>%
mutate( ACC = ACC ) %>%
mutate( F1 = F1 ) %>%
mutate( GINI = GINI ) %>%
mutate(Information = Information)
return( table_1_chisq_pvalue )
}\(~\)
\(~\)
\(~\)
21.10.1 Test lbxglu_value_chisq Function
Let’s test our function!
lbxglu_split_level[1] 135lbxglu_value_chisq(lbxglu_split_level, return_table=1)
Diabetes No Diabetes
lbxglu_over_value 101 31
lbxglu_under_value 68 926cm_1$table Reference
Prediction Diabetes No Diabetes
Diabetes 101 31
No Diabetes 68 926glimpse(lbxglu_value_chisq(lbxglu_split_level))Rows: 1
Columns: 12
$ lbxglu_value <dbl> 135
$ chisq_p_value <dbl> 2.944895e-97
$ TP <int> 101
$ FP <int> 31
$ FN <int> 68
$ TN <int> 926
$ PPV <dbl> 0.7651515
$ TPR <dbl> 0.5976331
$ ACC <dbl> 0.9120782
$ F1 <dbl> 0.6710963
$ GINI <dbl> 0.1546497
$ Information <dbl> 0.4099506\(~\)
\(~\)
\(~\)
21.11 Find Ranges
Now let’s find the ranges of values for which to apply our function:
range_lbxglu_by_diq010 <- train_set %>%
group_by(diq010) %>%
summarise(lbxglu_min = min(lbxglu,na.rm=TRUE) , lbxglu_max = max(lbxglu,na.rm=TRUE) )
range_lbxglu_by_diq010# A tibble: 2 × 3
diq010 lbxglu_min lbxglu_max
<fct> <dbl> <dbl>
1 Diabetes 50 479
2 No Diabetes 64 397my_min <- min(range_lbxglu_by_diq010$lbxglu_min) +1
my_min [1] 51# note anything less than `my_min` does not produce a 2x2 table:
lbxglu_value_chisq(my_min-1, return_table=1)
Diabetes No Diabetes
lbxglu_over_value 169 957lbxglu_value_chisq(my_min, return_table=1)
Diabetes No Diabetes
lbxglu_over_value 168 957
lbxglu_under_value 1 0my_max <- max(range_lbxglu_by_diq010$lbxglu_max)
my_max[1] 479# note anything more than `my_max` does not produce a 2x2 table:
lbxglu_value_chisq(my_max, return_table=1)
Diabetes No Diabetes
lbxglu_over_value 1 0
lbxglu_under_value 168 957lbxglu_value_chisq(my_max+1, return_table=1)
Diabetes No Diabetes
lbxglu_under_value 169 957# so the range of values are:
my_list <- my_min:my_max
my_list [1] 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68
[19] 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86
[37] 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104
[55] 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122
[73] 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140
[91] 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158
[109] 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176
[127] 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194
[145] 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212
[163] 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230
[181] 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248
[199] 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266
[217] 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284
[235] 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302
[253] 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320
[271] 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338
[289] 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356
[307] 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374
[325] 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392
[343] 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410
[361] 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428
[379] 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446
[397] 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464
[415] 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479\(~\)
\(~\)
\(~\)
21.12 Apply Function
Now we apply our function lbxglu_value_chisq to the range my_list
chi_square_lbxglu_value <- purrr::map_dfr(my_list, lbxglu_value_chisq)
glimpse(chi_square_lbxglu_value)Rows: 429
Columns: 12
$ lbxglu_value <int> 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, …
$ chisq_p_value <dbl> 0.3270151, 0.3270151, 0.3270151, 0.3270151, 0.3270151, 0…
$ TP <int> 168, 168, 168, 168, 168, 168, 168, 168, 168, 168, 168, 1…
$ FP <int> 957, 957, 957, 957, 957, 957, 957, 957, 957, 957, 957, 9…
$ FN <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ TN <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 2, 2, 2, 2,…
$ PPV <dbl> 0.1493333, 0.1493333, 0.1493333, 0.1493333, 0.1493333, 0…
$ TPR <dbl> 0.9940828, 0.9940828, 0.9940828, 0.9940828, 0.9940828, 0…
$ ACC <dbl> 0.1492007, 0.1492007, 0.1492007, 0.1492007, 0.1492007, 0…
$ F1 <dbl> 0.2596600, 0.2596600, 0.2596600, 0.2596600, 0.2596600, 0…
$ GINI <dbl> 0.2538401, 0.2538401, 0.2538401, 0.2538401, 0.2538401, 0…
$ Information <dbl> 0.6076293, 0.6076293, 0.6076293, 0.6076293, 0.6076293, 0…\(~\)
\(~\)
\(~\)
21.12.1 Sort Review Results
Let’s review the Results by sorting them with respect to different metrics:
chi_square_lbxglu_value %>% arrange(chisq_p_value)# A tibble: 429 × 12
lbxglu_value chisq_p_value TP FP FN TN PPV TPR ACC F1
<int> <dbl> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 135 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
2 136 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
3 141 3.60e-97 95 23 74 934 0.805 0.562 0.914 0.662
4 134 4.00e-97 103 34 66 923 0.752 0.609 0.911 0.673
5 133 1.26e-96 104 36 65 921 0.743 0.615 0.910 0.673
6 139 2.01e-96 96 25 73 932 0.793 0.568 0.913 0.662
7 145 2.68e-96 92 20 77 937 0.821 0.544 0.914 0.655
8 138 4.24e-96 98 28 71 929 0.778 0.580 0.912 0.664
9 140 4.29e-96 95 24 74 933 0.798 0.562 0.913 0.660
10 142 8.92e-96 94 23 75 934 0.803 0.556 0.913 0.657
# ℹ 419 more rows
# ℹ 2 more variables: GINI <dbl>, Information <dbl>chi_square_lbxglu_value %>% arrange(GINI)# A tibble: 429 × 12
lbxglu_value chisq_p_value TP FP FN TN PPV TPR ACC F1
<int> <dbl> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 135 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
2 136 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
3 141 3.60e-97 95 23 74 934 0.805 0.562 0.914 0.662
4 134 4.00e-97 103 34 66 923 0.752 0.609 0.911 0.673
5 133 1.26e-96 104 36 65 921 0.743 0.615 0.910 0.673
6 139 2.01e-96 96 25 73 932 0.793 0.568 0.913 0.662
7 145 2.68e-96 92 20 77 937 0.821 0.544 0.914 0.655
8 140 4.29e-96 95 24 74 933 0.798 0.562 0.913 0.660
9 138 4.24e-96 98 28 71 929 0.778 0.580 0.912 0.664
10 142 8.92e-96 94 23 75 934 0.803 0.556 0.913 0.657
# ℹ 419 more rows
# ℹ 2 more variables: GINI <dbl>, Information <dbl>chi_square_lbxglu_value %>% arrange(Information)# A tibble: 429 × 12
lbxglu_value chisq_p_value TP FP FN TN PPV TPR ACC F1
<int> <dbl> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 122 1.10e-90 120 71 49 886 0.628 0.710 0.893 0.667
2 134 4.00e-97 103 34 66 923 0.752 0.609 0.911 0.673
3 133 1.26e-96 104 36 65 921 0.743 0.615 0.910 0.673
4 135 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
5 136 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
6 130 7.29e-95 106 41 63 916 0.721 0.627 0.908 0.671
7 123 3.54e-90 117 66 52 891 0.639 0.692 0.895 0.665
8 121 4.26e-88 121 77 48 880 0.611 0.716 0.889 0.659
9 125 3.25e-91 114 59 55 898 0.659 0.675 0.899 0.667
10 131 2.21e-94 105 40 64 917 0.724 0.621 0.908 0.669
# ℹ 419 more rows
# ℹ 2 more variables: GINI <dbl>, Information <dbl>chi_square_lbxglu_value %>% arrange(-ACC)# A tibble: 429 × 12
lbxglu_value chisq_p_value TP FP FN TN PPV TPR ACC F1
<int> <dbl> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 141 3.60e-97 95 23 74 934 0.805 0.562 0.914 0.662
2 145 2.68e-96 92 20 77 937 0.821 0.544 0.914 0.655
3 139 2.01e-96 96 25 73 932 0.793 0.568 0.913 0.662
4 140 4.29e-96 95 24 74 933 0.798 0.562 0.913 0.660
5 142 8.92e-96 94 23 75 934 0.803 0.556 0.913 0.657
6 146 6.67e-95 91 20 78 937 0.820 0.538 0.913 0.65
7 147 1.22e-94 90 19 79 938 0.826 0.533 0.913 0.647
8 149 3.68e-94 88 17 81 940 0.838 0.521 0.913 0.642
9 152 1.43e-93 85 14 84 943 0.859 0.503 0.913 0.634
10 153 1.43e-93 85 14 84 943 0.859 0.503 0.913 0.634
# ℹ 419 more rows
# ℹ 2 more variables: GINI <dbl>, Information <dbl>chi_square_lbxglu_value %>% arrange(-PPV)# A tibble: 429 × 12
lbxglu_value chisq_p_value TP FP FN TN PPV TPR ACC F1
<int> <dbl> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 398 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
2 399 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
3 400 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
4 401 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
5 402 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
6 403 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
7 404 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
8 405 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
9 406 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
10 407 0.00000253 5 0 164 957 1 0.0296 0.854 0.0575
# ℹ 419 more rows
# ℹ 2 more variables: GINI <dbl>, Information <dbl>chi_square_lbxglu_value %>% arrange(-F1)# A tibble: 429 × 12
lbxglu_value chisq_p_value TP FP FN TN PPV TPR ACC F1
<int> <dbl> <int> <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
1 134 4.00e-97 103 34 66 923 0.752 0.609 0.911 0.673
2 133 1.26e-96 104 36 65 921 0.743 0.615 0.910 0.673
3 135 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
4 136 2.94e-97 101 31 68 926 0.765 0.598 0.912 0.671
5 130 7.29e-95 106 41 63 916 0.721 0.627 0.908 0.671
6 131 2.21e-94 105 40 64 917 0.724 0.621 0.908 0.669
7 122 1.10e-90 120 71 49 886 0.628 0.710 0.893 0.667
8 125 3.25e-91 114 59 55 898 0.659 0.675 0.899 0.667
9 129 3.82e-93 106 43 63 914 0.711 0.627 0.906 0.667
10 132 6.57e-94 104 39 65 918 0.727 0.615 0.908 0.667
# ℹ 419 more rows
# ℹ 2 more variables: GINI <dbl>, Information <dbl>\(~\)
\(~\)
\(~\)
21.13 Plot Results
Let’s Plot Our Results
glimpse(chi_square_lbxglu_value) Rows: 429
Columns: 12
$ lbxglu_value <int> 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, …
$ chisq_p_value <dbl> 0.3270151, 0.3270151, 0.3270151, 0.3270151, 0.3270151, 0…
$ TP <int> 168, 168, 168, 168, 168, 168, 168, 168, 168, 168, 168, 1…
$ FP <int> 957, 957, 957, 957, 957, 957, 957, 957, 957, 957, 957, 9…
$ FN <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ TN <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 2, 2, 2, 2,…
$ PPV <dbl> 0.1493333, 0.1493333, 0.1493333, 0.1493333, 0.1493333, 0…
$ TPR <dbl> 0.9940828, 0.9940828, 0.9940828, 0.9940828, 0.9940828, 0…
$ ACC <dbl> 0.1492007, 0.1492007, 0.1492007, 0.1492007, 0.1492007, 0…
$ F1 <dbl> 0.2596600, 0.2596600, 0.2596600, 0.2596600, 0.2596600, 0…
$ GINI <dbl> 0.2538401, 0.2538401, 0.2538401, 0.2538401, 0.2538401, 0…
$ Information <dbl> 0.6076293, 0.6076293, 0.6076293, 0.6076293, 0.6076293, 0…chi_square_lbxglu_value.ggplot_data <- chi_square_lbxglu_value %>%
select(lbxglu_value, chisq_p_value, PPV, TPR, ACC, F1, GINI, Information) %>%
gather(-lbxglu_value, key="stat_test", value="Value")
glimpse(chi_square_lbxglu_value.ggplot_data)Rows: 3,003
Columns: 3
$ lbxglu_value <int> 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 6…
$ stat_test <chr> "chisq_p_value", "chisq_p_value", "chisq_p_value", "chisq…
$ Value <dbl> 0.3270151, 0.3270151, 0.3270151, 0.3270151, 0.3270151, 0.…library('ggplot2')
plot_1 <- chi_square_lbxglu_value.ggplot_data %>%
ggplot(aes(x=lbxglu_value, y=Value, color=stat_test)) +
geom_point()
plot_1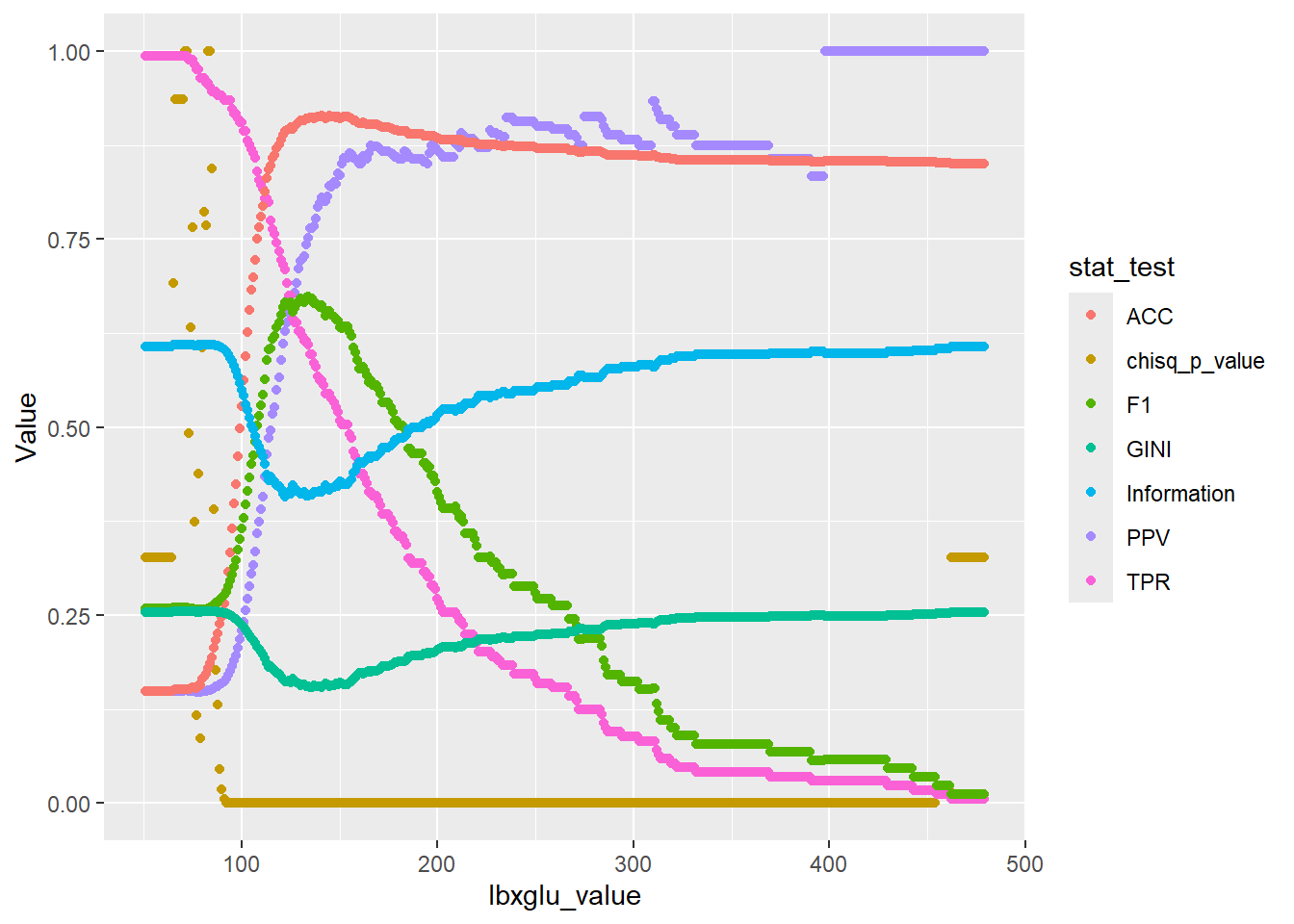
\(~\)
\(~\)
Now let’s add in the lbxglu_split_level from the decision tree:
plot_1 + geom_vline(xintercept = lbxglu_split_level)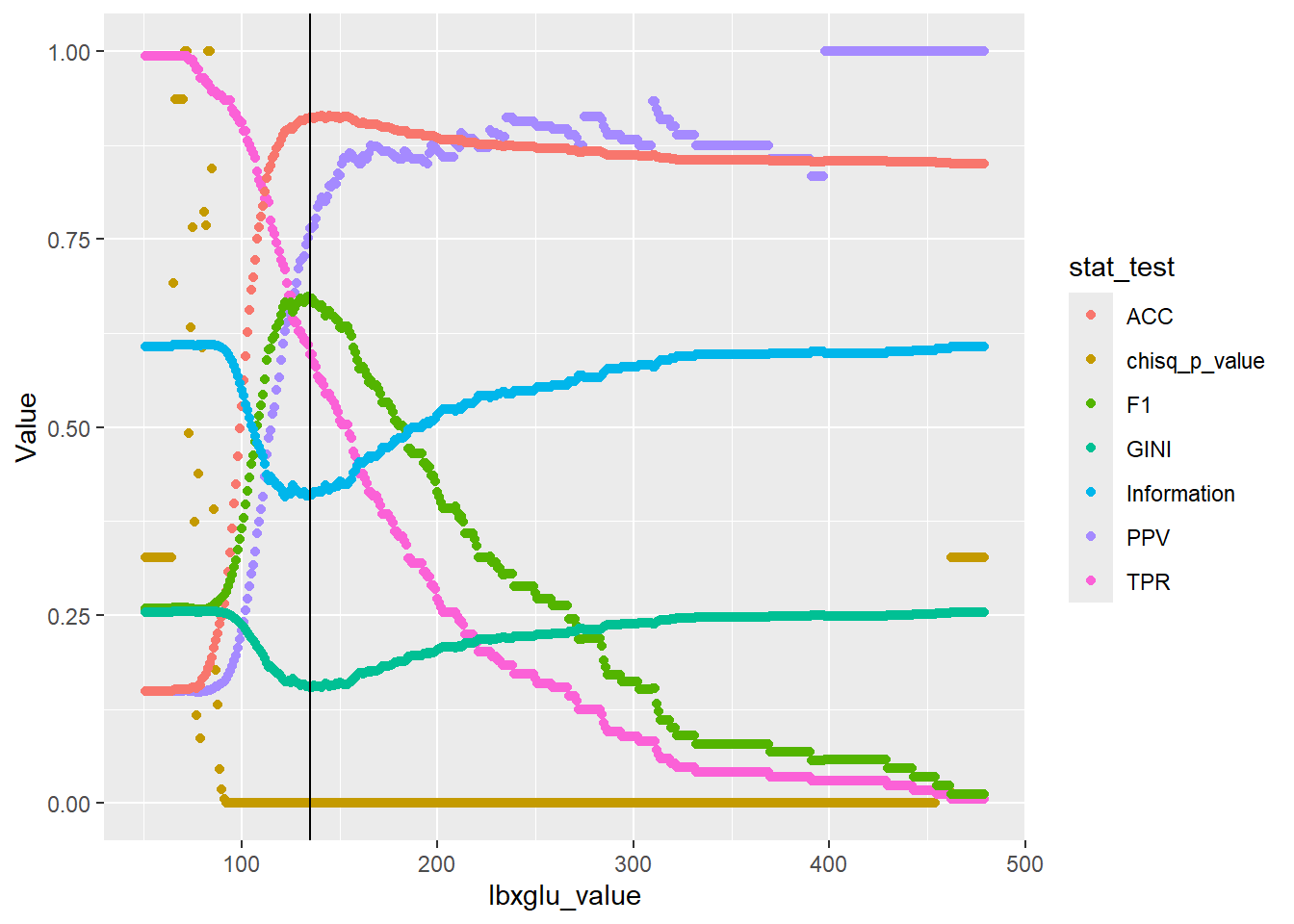
\(~\)
While some metrics such as PPV continue to impove, we can see that the F1 score is maximized around lbxglu_split_level.
Typically, when fitting a decision tree the GINI or Information is used to determine the splits and the order of the splits.
Hopefully, this gives some indication of how why split value lbxglu_split_level is equal to 135.
\(~\)
\(~\)
\(~\)
\(~\)
22 Score The Test Data
Now that we have a better understanding of what the decision tree model is doing, we will score the test data:
test <- diab_pop.no_na_vals.test
test.prune.y_hat_probs <- predict(tree_prune, test)
test.prune.y_hat_class <- predict(tree_prune, test, type ="class")
test.prune_scored <- as_tibble(cbind(test, test.prune.y_hat_probs, test.prune.y_hat_class))
glimpse(test.prune_scored)Rows: 750
Columns: 13
$ seqn <dbl> 83734, 83737, 83757, 83761, 83789, 83820, 83822…
$ riagendr <fct> Male, Female, Female, Female, Male, Male, Femal…
$ ridageyr <dbl> 78, 72, 57, 24, 66, 70, 20, 29, 69, 71, 37, 49,…
$ ridreth1 <fct> Non-Hispanic White, MexicanAmerican, Other Hisp…
$ dmdeduc2 <fct> High school graduate/GED, Grades 9-11th, Less t…
$ dmdmartl <fct> Married, Separated, Separated, Never married, L…
$ indhhin2 <fct> "$20,000-$24,999", "$75,000-$99,999", "$20,000-…
$ bmxbmi <dbl> 28.8, 28.6, 35.4, 25.3, 34.0, 27.0, 22.2, 29.7,…
$ diq010 <fct> Diabetes, No Diabetes, Diabetes, No Diabetes, N…
$ lbxglu <dbl> 84, 107, 398, 95, 113, 94, 80, 102, 105, 76, 79…
$ Diabetes <dbl> 0.06841046, 0.06841046, 0.76515152, 0.06841046,…
$ `No Diabetes` <dbl> 0.9315895, 0.9315895, 0.2348485, 0.9315895, 0.9…
$ test.prune.y_hat_class <fct> No Diabetes, No Diabetes, Diabetes, No Diabetes…prune_cm_test <- confusionMatrix(data = test.prune_scored$test.prune.y_hat_class,
reference = test.prune_scored$diq010,
positive = 'Diabetes',
mode = "everything")
prune_cm_testConfusion Matrix and Statistics
Reference
Prediction Diabetes No Diabetes
Diabetes 63 21
No Diabetes 49 617
Accuracy : 0.9067
95% CI : (0.8836, 0.9265)
No Information Rate : 0.8507
P-Value [Acc > NIR] : 3.387e-06
Kappa : 0.5904
Mcnemar's Test P-Value : 0.00125
Sensitivity : 0.5625
Specificity : 0.9671
Pos Pred Value : 0.7500
Neg Pred Value : 0.9264
Precision : 0.7500
Recall : 0.5625
F1 : 0.6429
Prevalence : 0.1493
Detection Rate : 0.0840
Detection Prevalence : 0.1120
Balanced Accuracy : 0.7648
'Positive' Class : Diabetes
\(~\)
\(~\)
22.1 Use yardstick for Model Metrics
install_if_not('yardstick')[1] "the package 'yardstick' is already installed"library('yardstick')
Attaching package: 'yardstick'The following objects are masked from 'package:caret':
precision, recall, sensitivity, specificityThe following object is masked from 'package:readr':
speccm_2 <- test.prune_scored %>%
conf_mat(truth = diq010, estimate = test.prune.y_hat_class )
summary(cm_2)# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.907
2 kap binary 0.590
3 sens binary 0.562
4 spec binary 0.967
5 ppv binary 0.75
6 npv binary 0.926
7 mcc binary 0.599
8 j_index binary 0.530
9 bal_accuracy binary 0.765
10 detection_prevalence binary 0.112
11 precision binary 0.75
12 recall binary 0.562
13 f_meas binary 0.643accuracy_val <- (summary(cm_2) %>% filter(.metric == 'accuracy'))$.estimate
accuracy_val[1] 0.9066667autoplot(cm_2)
autoplot(cm_2, type = "heatmap")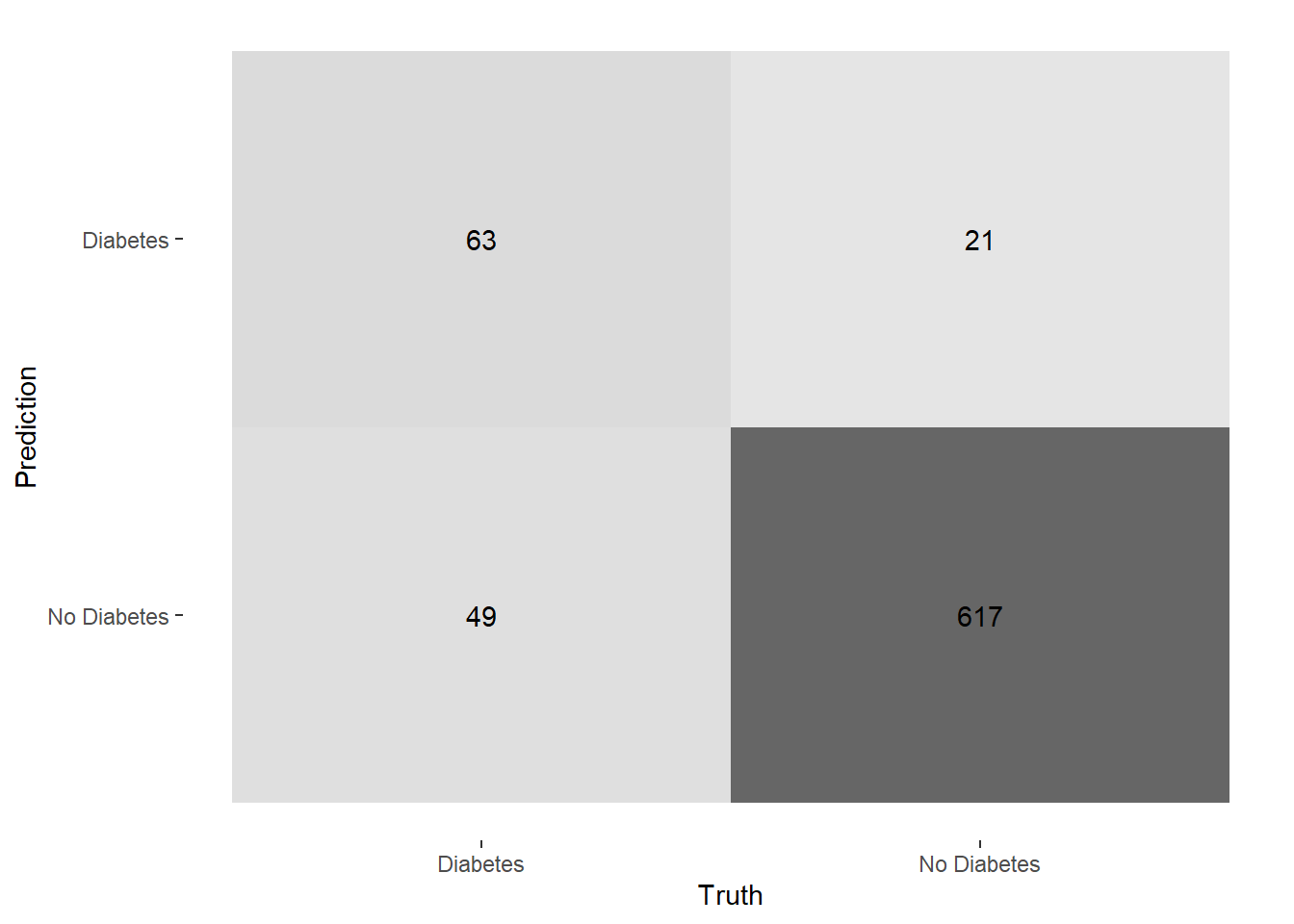
str(cm_2)List of 1
$ table: 'table' num [1:2, 1:2] 63 49 21 617
..- attr(*, "dimnames")=List of 2
.. ..$ Prediction: chr [1:2] "Diabetes" "No Diabetes"
.. ..$ Truth : chr [1:2] "Diabetes" "No Diabetes"
- attr(*, "class")= chr "conf_mat"gplots::balloonplot(cm_2$table,
main ="Balloon Plot Confusion Matrix for Pruned Model \n Area is proportional to Freq.")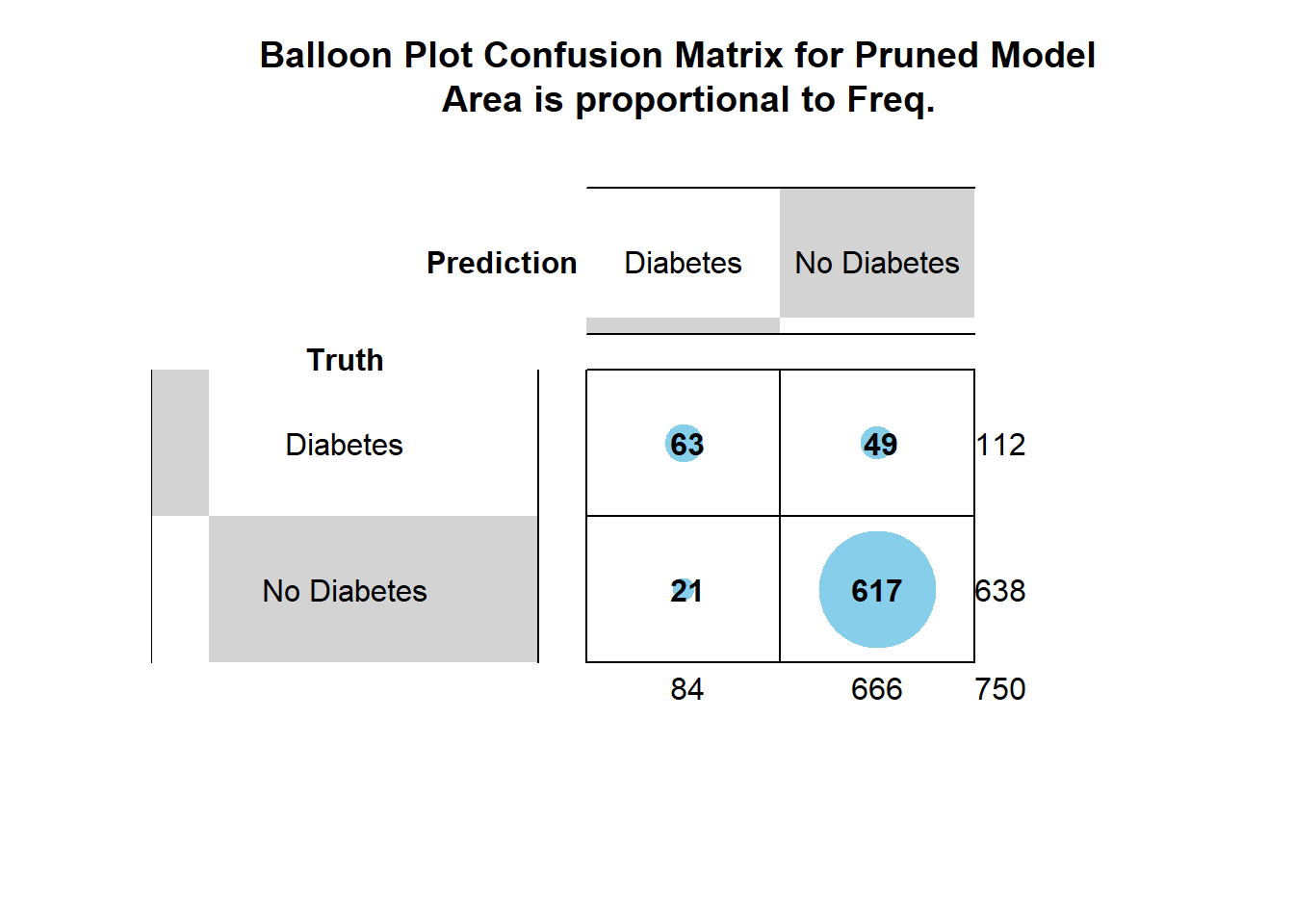
\(~\)
\(~\)
\(~\)
22.2 ROC Curve
metrics.prune <- test.prune_scored %>%
metrics(truth=diq010, test.prune.y_hat_class)
metrics.prune# A tibble: 2 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.907
2 kap binary 0.590roc_auc.prune <- test.prune_scored %>%
roc_auc(truth=diq010, Diabetes)
roc_auc.prune# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.765test_prune_roc <- test.prune_scored %>%
roc_curve(truth=diq010, Diabetes)
autoplot(test_prune_roc)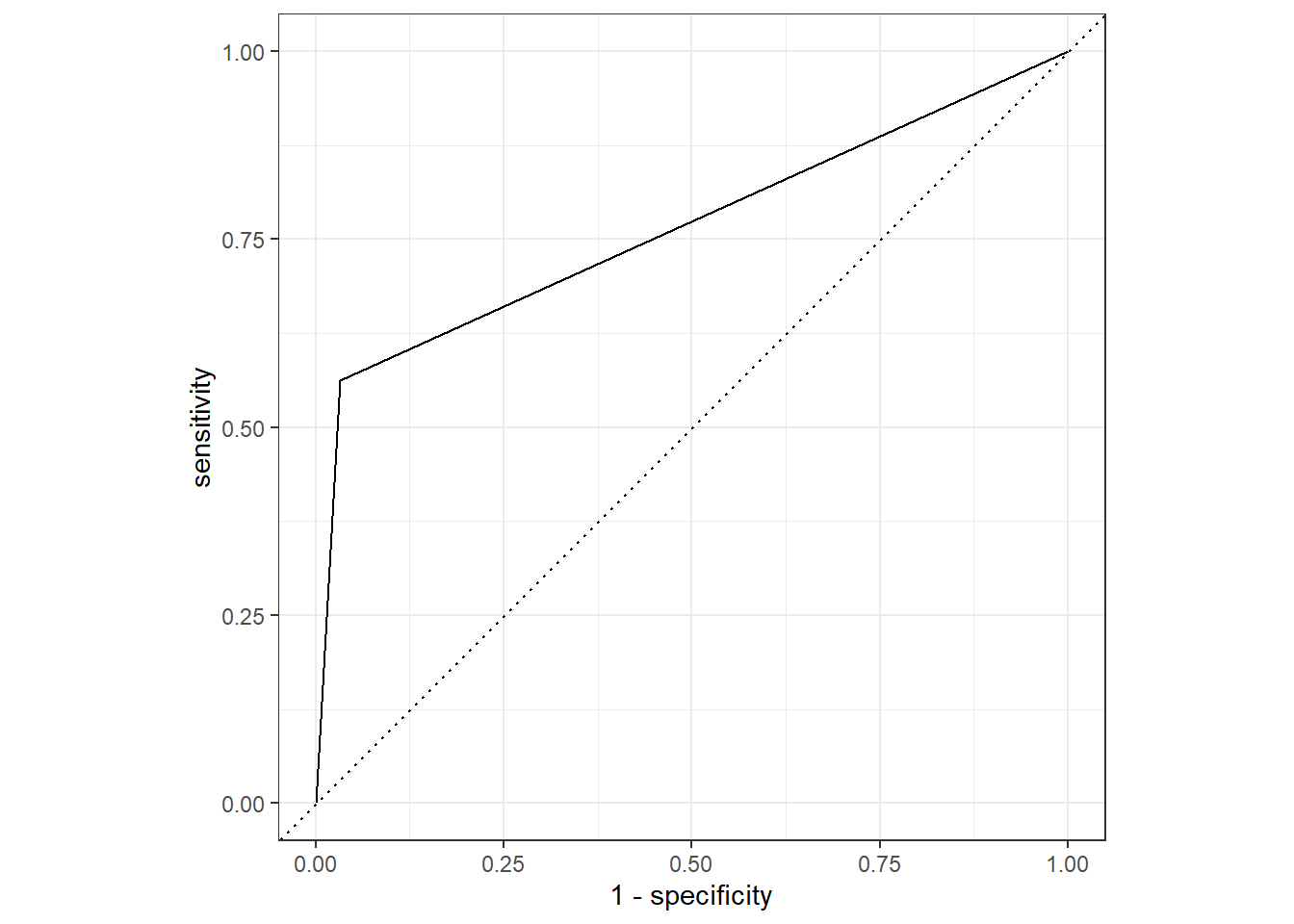
plot_1 <- test_prune_roc %>%
ggplot(aes(x = 1 - specificity, y = sensitivity)) +
geom_path() +
geom_abline(lty = 3) +
coord_equal() +
theme_bw()
plot_1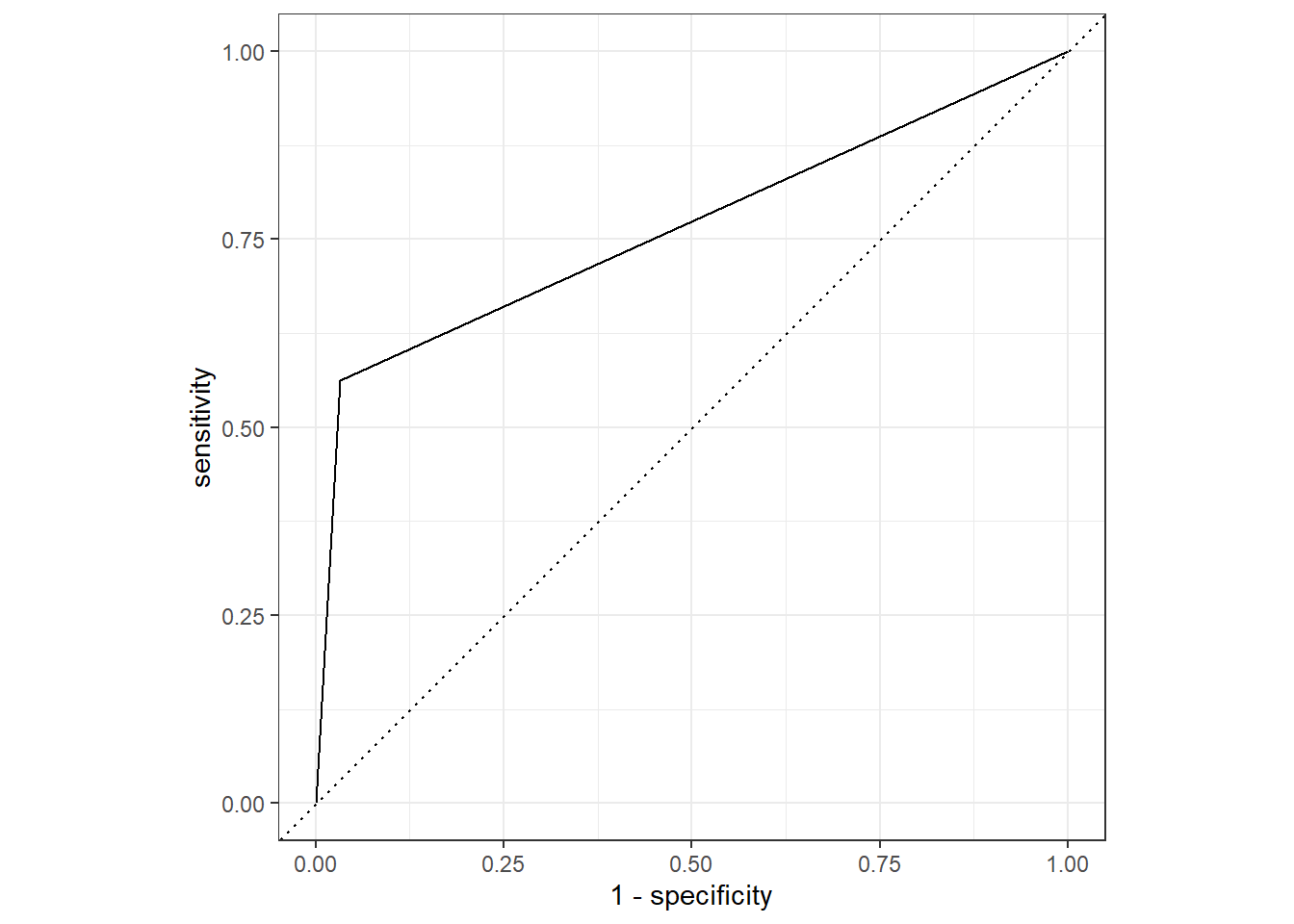
autoplot(test_prune_roc) +
labs( title = "ROC Curve - Pruned Model",
caption = paste0("Area Under ROC Curve : ", round(roc_auc.prune$.estimate,3) ) ) +
theme( plot.title = element_text(size = 18) ,
plot.caption = element_text(size = 12))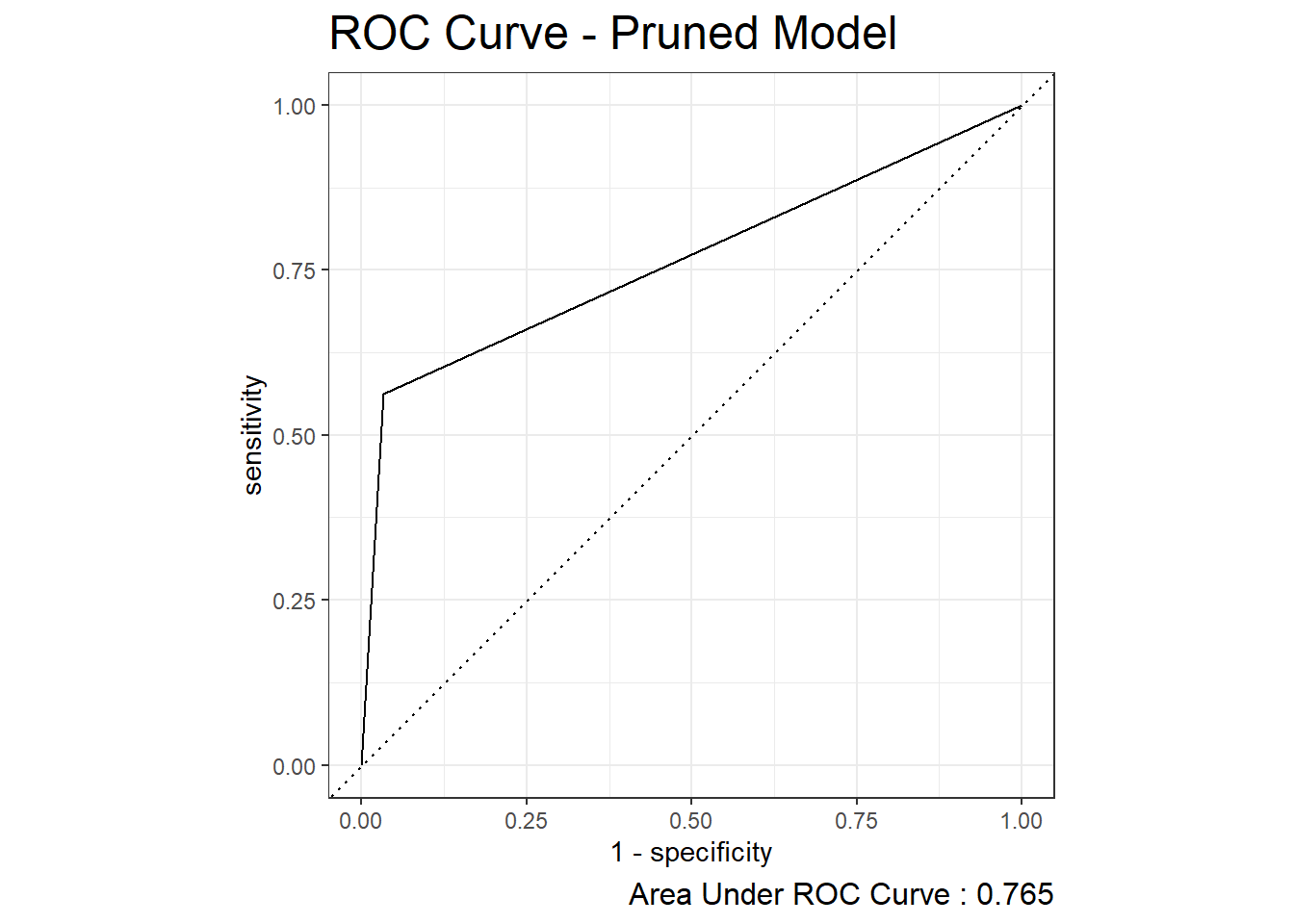
\(~\)
\(~\)
\(~\)
22.3 Precision Recall Curve
test_prune_pr_auc <- test.prune_scored %>%
pr_auc(truth=diq010, Diabetes)
test_prune_pr_auc# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 pr_auc binary 0.689test_prune_precision_recall <- test.prune_scored %>%
pr_curve(truth=diq010, Diabetes)
autoplot(test_prune_precision_recall) +
labs( title = "Precision Recall Curve - Pruned Model",
caption = paste0("Area Under Precision Recall Curve : ", round(test_prune_pr_auc$.estimate,3) ) ) +
theme( plot.title = element_text(size = 18) ,
plot.caption = element_text(size = 12))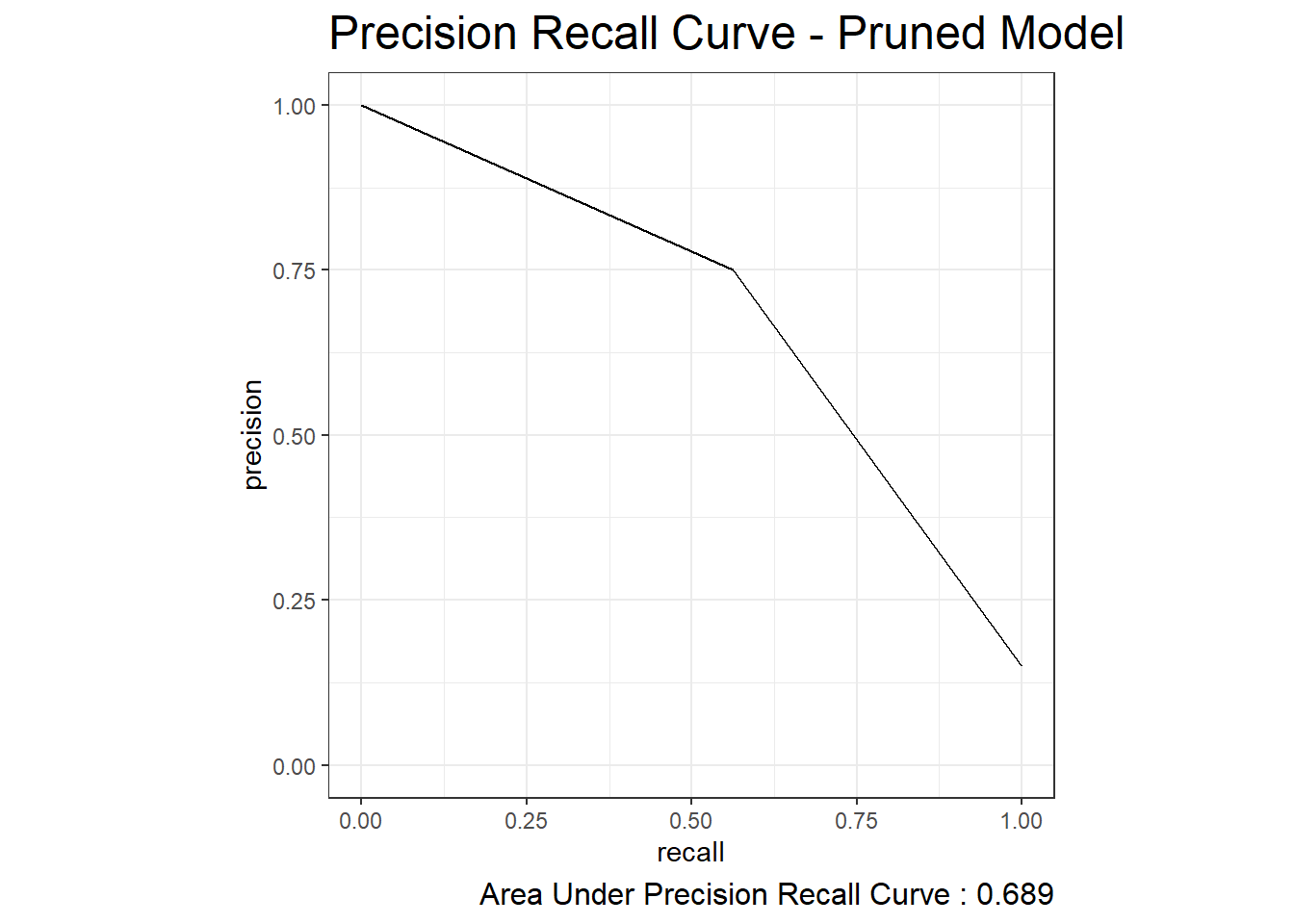
\(~\)
\(~\)
\(~\)
22.4 Lift Curve
test_prune_lift <- test.prune_scored %>%
lift_curve(truth=diq010, Diabetes)
autoplot(test_prune_lift) +
labs( title = "Lift Curve - Pruned Model") +
theme(plot.title = element_text(size = 18))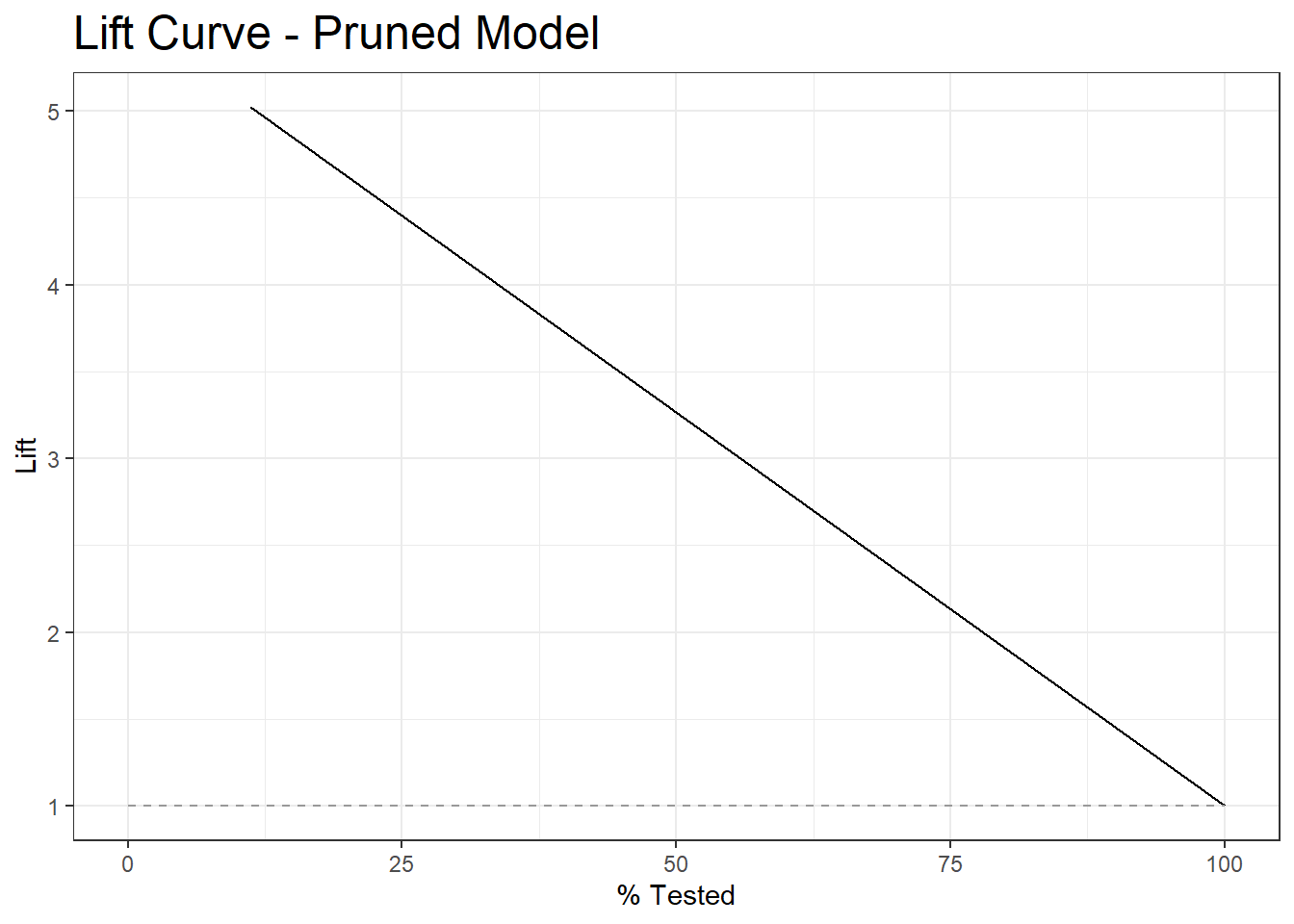
\(~\)
\(~\)
\(~\)
22.5 Gain Curve
test_prune_gain <- test.prune_scored %>%
gain_curve(truth=diq010, Diabetes)
autoplot(test_prune_gain) +
labs( title = "Gain Curve - Pruned Model") +
theme(plot.title = element_text(size = 18))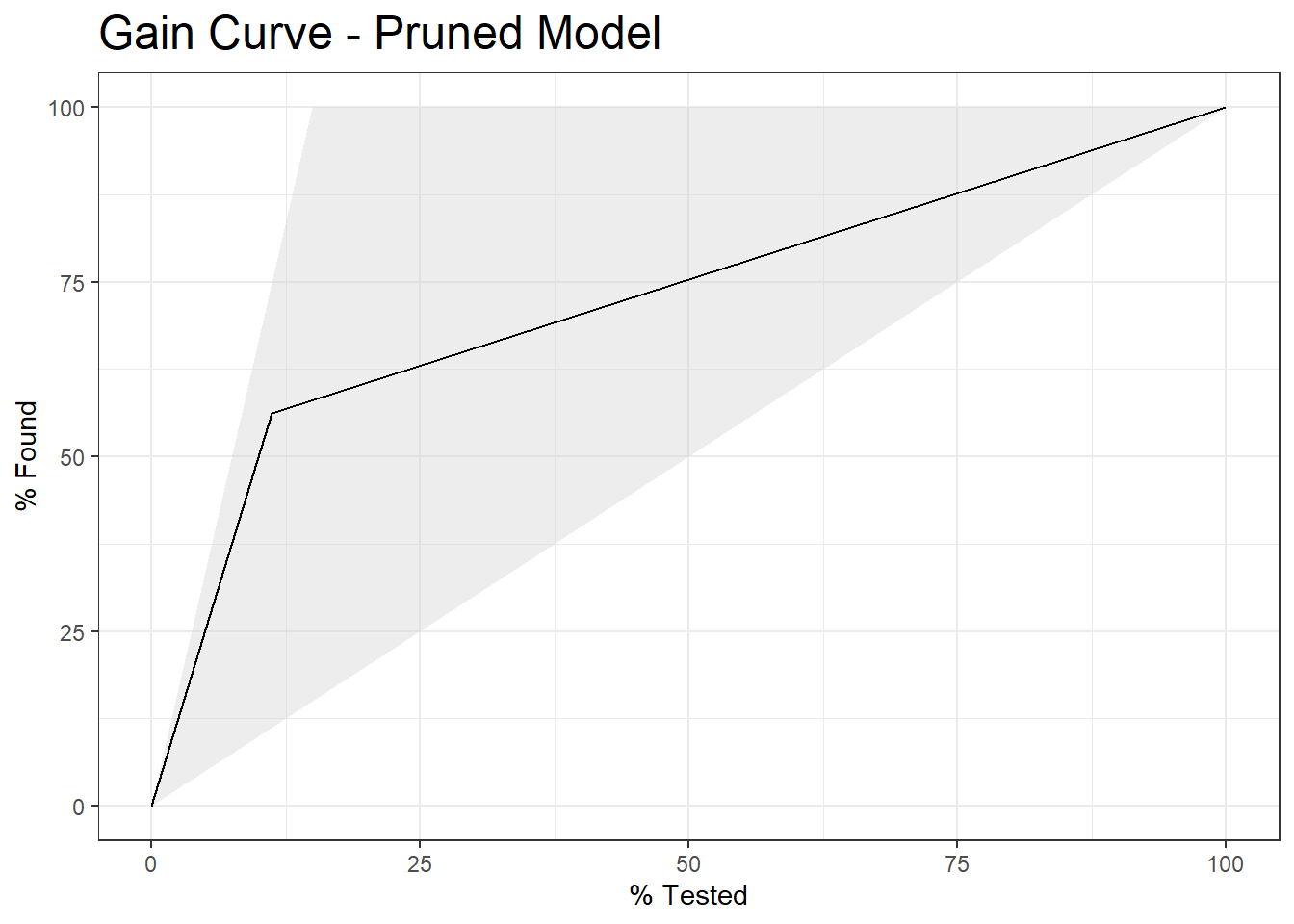
\(~\)
\(~\)
\(~\)
\(~\)
22.6 Comparing Models
A common task will be to compare the effectiveness of two models.
In this case, we will compare our pruned model to our origional model.
# pruned model
glimpse(test.prune_scored)Rows: 750
Columns: 13
$ seqn <dbl> 83734, 83737, 83757, 83761, 83789, 83820, 83822…
$ riagendr <fct> Male, Female, Female, Female, Male, Male, Femal…
$ ridageyr <dbl> 78, 72, 57, 24, 66, 70, 20, 29, 69, 71, 37, 49,…
$ ridreth1 <fct> Non-Hispanic White, MexicanAmerican, Other Hisp…
$ dmdeduc2 <fct> High school graduate/GED, Grades 9-11th, Less t…
$ dmdmartl <fct> Married, Separated, Separated, Never married, L…
$ indhhin2 <fct> "$20,000-$24,999", "$75,000-$99,999", "$20,000-…
$ bmxbmi <dbl> 28.8, 28.6, 35.4, 25.3, 34.0, 27.0, 22.2, 29.7,…
$ diq010 <fct> Diabetes, No Diabetes, Diabetes, No Diabetes, N…
$ lbxglu <dbl> 84, 107, 398, 95, 113, 94, 80, 102, 105, 76, 79…
$ Diabetes <dbl> 0.06841046, 0.06841046, 0.76515152, 0.06841046,…
$ `No Diabetes` <dbl> 0.9315895, 0.9315895, 0.2348485, 0.9315895, 0.9…
$ test.prune.y_hat_class <fct> No Diabetes, No Diabetes, Diabetes, No Diabetes…test.prune_scored_sel <- test.prune_scored %>%
select(seqn,diq010, Diabetes, test.prune.y_hat_class) %>%
rename(pred_prob = Diabetes) %>%
rename(pred_class = test.prune.y_hat_class) %>%
mutate(model_type = 'prune')
# Score the Original Model on Test Data
test.y_hat_probs <- predict(tree_1, test)
test.y_hat_class <- predict(tree_1, test, type ="class")
test.scored <- as_tibble(cbind(test, test.y_hat_probs, test.y_hat_class))
test.scored_sel <- test.scored %>%
select(seqn,diq010, Diabetes, test.y_hat_class) %>%
rename(pred_prob = Diabetes) %>%
rename(pred_class = test.y_hat_class) %>%
mutate(model_type = 'not_pruned')
stacked_dfs <- rbind(test.prune_scored_sel, test.scored_sel)
glimpse(stacked_dfs)Rows: 1,500
Columns: 5
$ seqn <dbl> 83734, 83737, 83757, 83761, 83789, 83820, 83822, 83823, 838…
$ diq010 <fct> Diabetes, No Diabetes, Diabetes, No Diabetes, No Diabetes, …
$ pred_prob <dbl> 0.06841046, 0.06841046, 0.76515152, 0.06841046, 0.06841046,…
$ pred_class <fct> No Diabetes, No Diabetes, Diabetes, No Diabetes, No Diabete…
$ model_type <chr> "prune", "prune", "prune", "prune", "prune", "prune", "prun…22.7 Compare Model Metrics
cm_compare <- stacked_dfs %>%
group_by(model_type) %>%
conf_mat(truth = diq010, estimate = pred_class )
cm_compare# A tibble: 2 × 2
model_type conf_mat
<chr> <list>
1 not_pruned <conf_mat>
2 prune <conf_mat>cm_compare$conf_mat[[1]]
Truth
Prediction Diabetes No Diabetes
Diabetes 60 32
No Diabetes 52 606
[[2]]
Truth
Prediction Diabetes No Diabetes
Diabetes 63 21
No Diabetes 49 617(cm_compare %>% filter(model_type == 'prune'))$conf_mat[[1]]
Truth
Prediction Diabetes No Diabetes
Diabetes 63 21
No Diabetes 49 617prune_cm <- (cm_compare %>% filter(model_type == 'prune'))$conf_mat[[1]]
prune_cm Truth
Prediction Diabetes No Diabetes
Diabetes 63 21
No Diabetes 49 617summary(prune_cm)# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.907
2 kap binary 0.590
3 sens binary 0.562
4 spec binary 0.967
5 ppv binary 0.75
6 npv binary 0.926
7 mcc binary 0.599
8 j_index binary 0.530
9 bal_accuracy binary 0.765
10 detection_prevalence binary 0.112
11 precision binary 0.75
12 recall binary 0.562
13 f_meas binary 0.643not_pruned_cm <- (cm_compare %>% filter(model_type == 'not_pruned'))$conf_mat[[1]]
not_pruned_cm Truth
Prediction Diabetes No Diabetes
Diabetes 60 32
No Diabetes 52 606summary(not_pruned_cm)# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.888
2 kap binary 0.524
3 sens binary 0.536
4 spec binary 0.950
5 ppv binary 0.652
6 npv binary 0.921
7 mcc binary 0.528
8 j_index binary 0.486
9 bal_accuracy binary 0.743
10 detection_prevalence binary 0.123
11 precision binary 0.652
12 recall binary 0.536
13 f_meas binary 0.588compared_cm_stats <- summary(not_pruned_cm) %>%
left_join(summary(prune_cm),
by=c(".metric",".estimator"),
suffix = c("","_prune")) %>%
gather(-.metric,-.estimator, key="prune", value= Value)
compared_cm_stats# A tibble: 26 × 4
.metric .estimator prune Value
<chr> <chr> <chr> <dbl>
1 accuracy binary .estimate 0.888
2 kap binary .estimate 0.524
3 sens binary .estimate 0.536
4 spec binary .estimate 0.950
5 ppv binary .estimate 0.652
6 npv binary .estimate 0.921
7 mcc binary .estimate 0.528
8 j_index binary .estimate 0.486
9 bal_accuracy binary .estimate 0.743
10 detection_prevalence binary .estimate 0.123
# ℹ 16 more rowsggplot(compared_cm_stats, aes(.metric, Value, fill = prune)) +
geom_bar(stat="identity", position=position_dodge()) +
coord_flip() 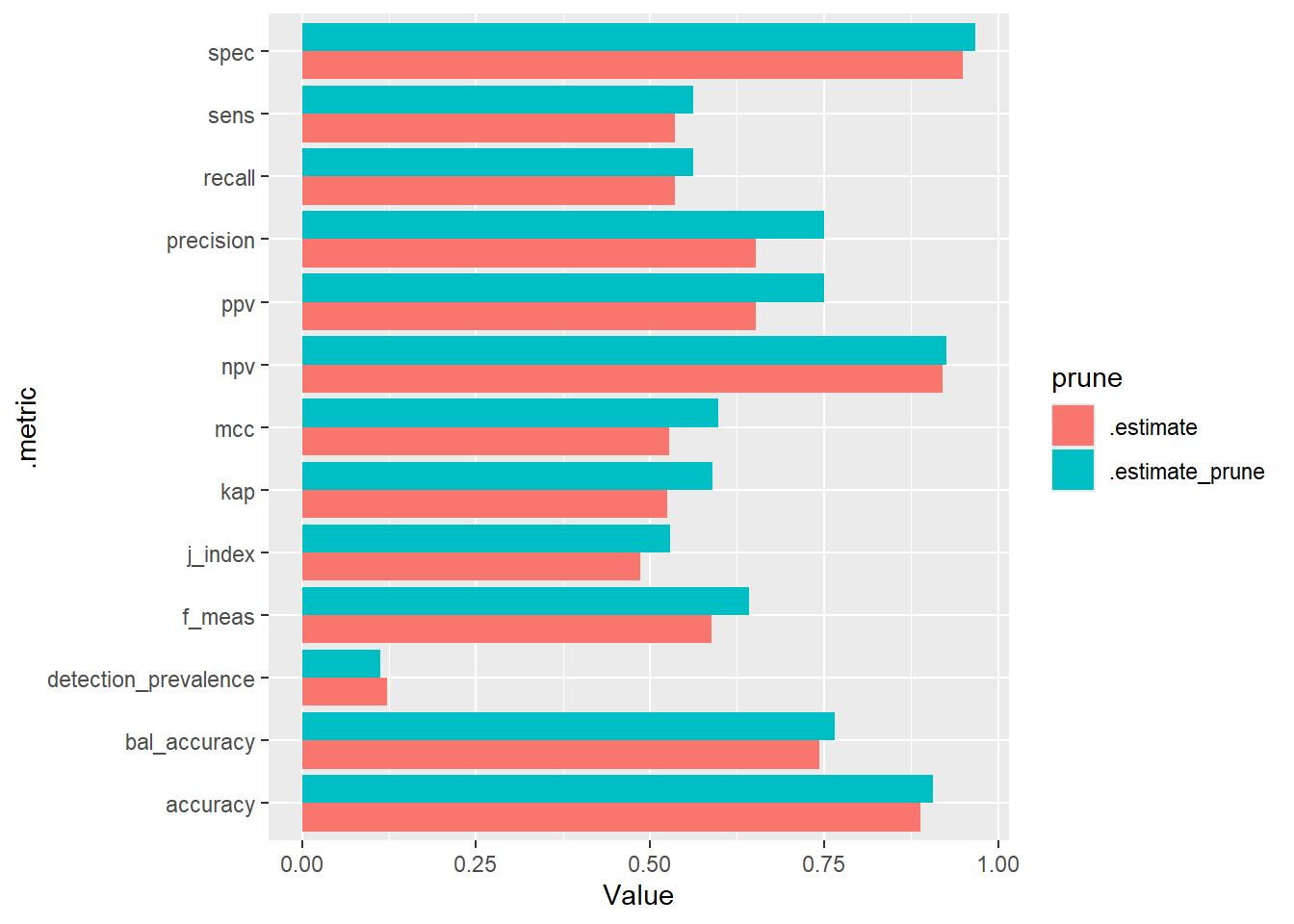
22.8 Compare ROC Curves
roc_auc.compare <- stacked_dfs %>%
group_by(model_type) %>%
roc_auc(truth=diq010, pred_prob)
roc_auc.compare# A tibble: 2 × 4
model_type .metric .estimator .estimate
<chr> <chr> <chr> <dbl>
1 not_pruned roc_auc binary 0.780
2 prune roc_auc binary 0.765roc_auc.compare2 <- roc_auc.compare %>%
select(model_type, .estimate) %>%
spread(key='model_type',value='.estimate')
roc_auc.compare2# A tibble: 1 × 2
not_pruned prune
<dbl> <dbl>
1 0.780 0.765test_compare_roc <- stacked_dfs %>%
group_by(model_type) %>%
roc_curve(truth=diq010, pred_prob)
autoplot(test_compare_roc) +
labs( caption = paste0("ROC_AUC NOT PRUNED: ", round(roc_auc.compare2$not_pruned,3) ,
"\nROC_AUC PRUNED: ", round(roc_auc.compare2$prune,3) ) )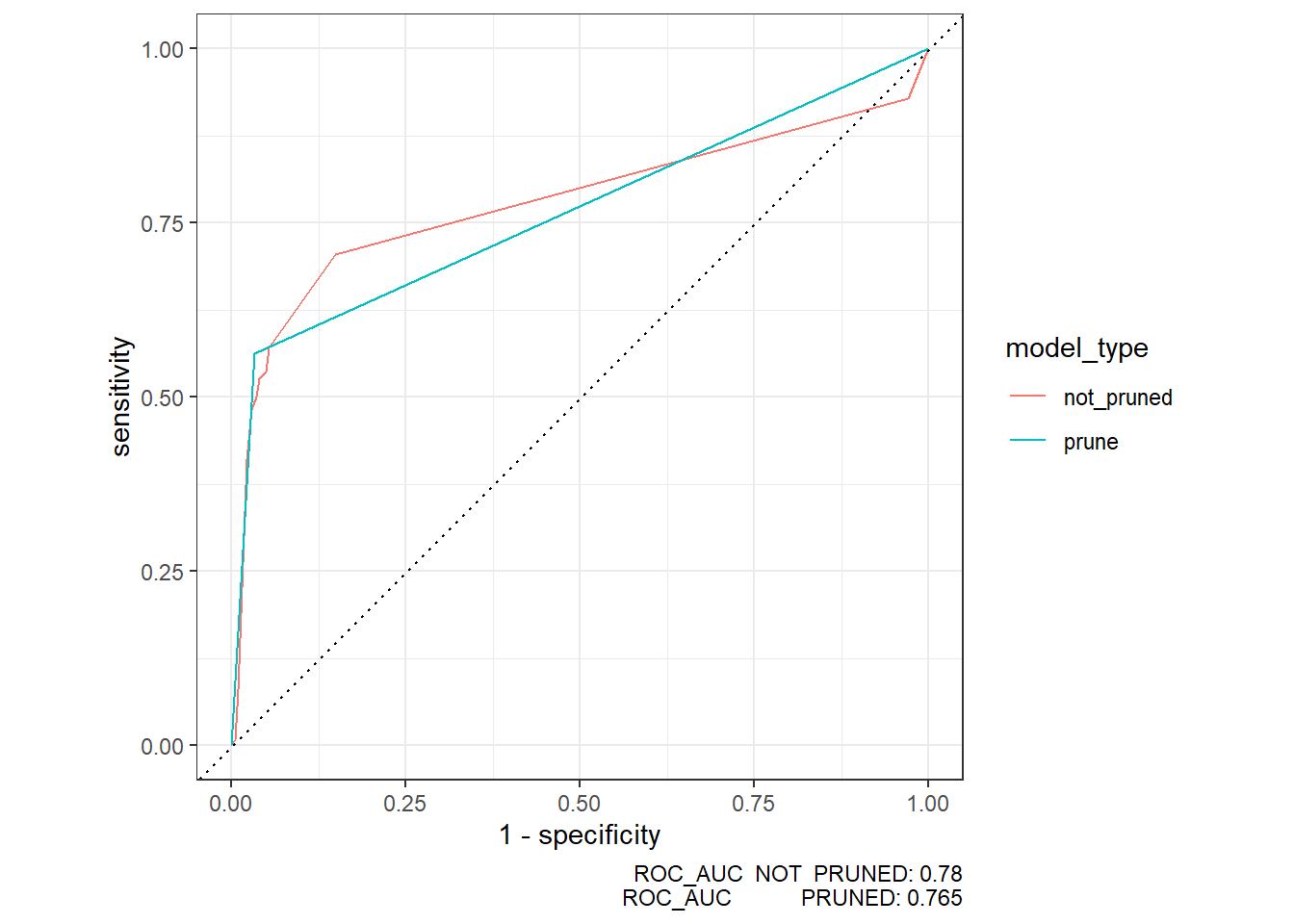
\(~\)
\(~\)
\(~\)
\(~\)
22.9 Compare Model Metrics - More Groups
diab_pop.test.stacked_dfs <- stacked_dfs %>%
left_join(diab_pop.no_na_vals.test, by = c("seqn", "diq010"))
rpart.plot(tree_1)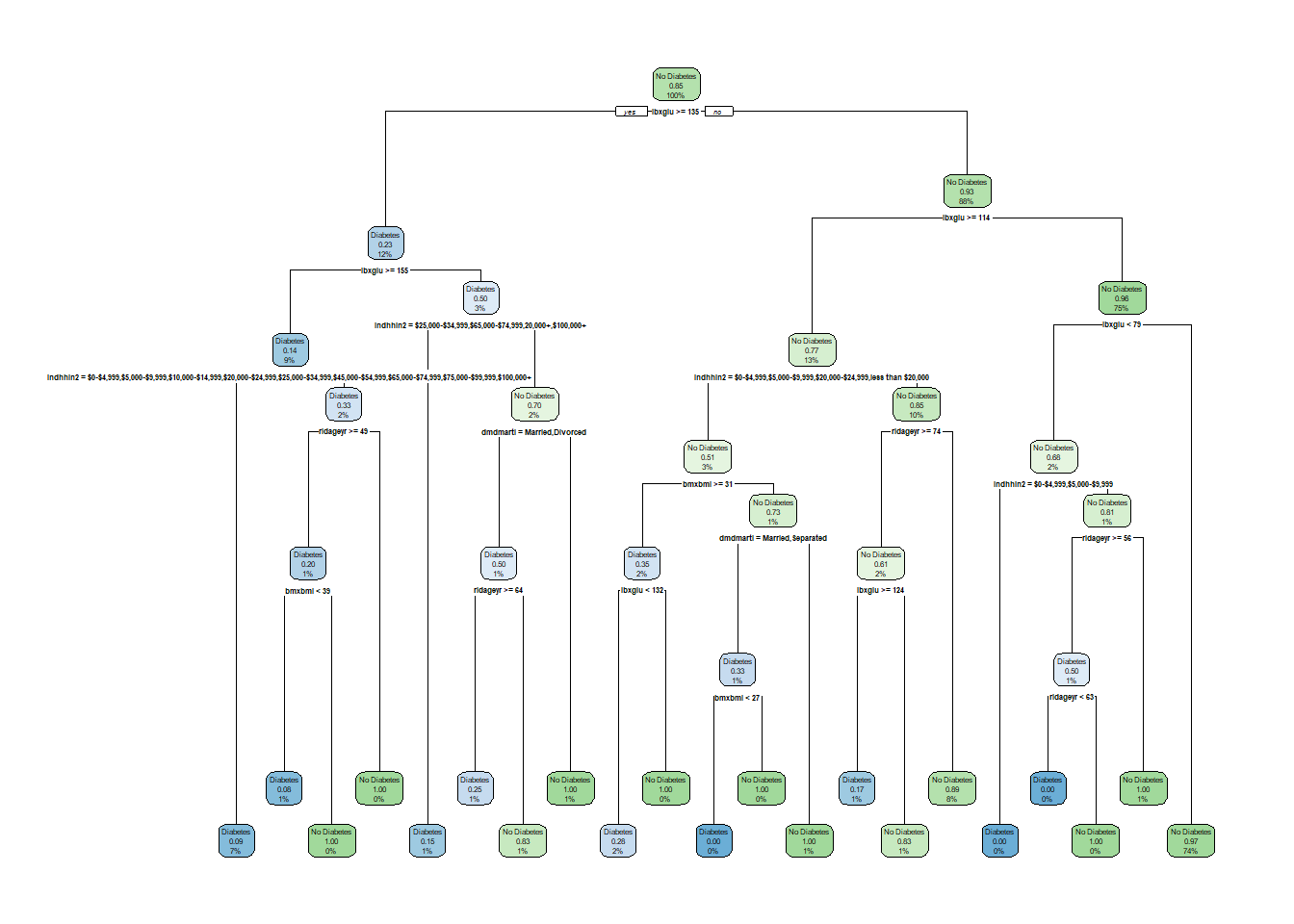
head(tree_1$splits,20) count ncat improve index adj
lbxglu 1126 1 113.1344112 135.00 0.00000000
ridageyr 1126 1 22.8255083 48.50 0.00000000
bmxbmi 1126 1 7.6602898 27.65 0.00000000
dmdeduc2 1126 5 5.7790434 1.00 0.00000000
dmdmartl 1126 6 5.7449142 2.00 0.00000000
lbxglu 132 1 6.9602273 154.50 0.00000000
bmxbmi 132 1 4.1704107 25.25 0.00000000
indhhin2 132 14 2.8812328 3.00 0.00000000
ridageyr 132 1 2.3778555 27.50 0.00000000
dmdmartl 132 6 1.8782314 4.00 0.00000000
ridageyr 0 1 0.7424242 27.50 0.05555556
bmxbmi 0 1 0.7424242 20.85 0.05555556
indhhin2 0 14 0.7348485 5.00 0.02777778
indhhin2 96 14 1.7355769 6.00 0.00000000
bmxbmi 96 -1 1.0405963 37.20 0.00000000
ridreth1 96 5 0.5720238 7.00 0.00000000
dmdmartl 96 6 0.5628608 8.00 0.00000000
ridageyr 96 1 0.5208333 61.50 0.00000000
bmxbmi 0 1 0.8333333 21.35 0.11111111
lbxglu 0 -1 0.8333333 393.50 0.11111111# let's use ridreth1
cm_compare_groups <- diab_pop.test.stacked_dfs %>%
group_by(model_type,ridreth1) %>%
conf_mat(truth = diq010, estimate = pred_class ) %>%
ungroup()
cm_compare_groups# A tibble: 10 × 3
model_type ridreth1 conf_mat
<chr> <fct> <list>
1 not_pruned MexicanAmerican <conf_mat>
2 not_pruned Other Hispanic <conf_mat>
3 not_pruned Non-Hispanic White <conf_mat>
4 not_pruned Non-Hispanic Black <conf_mat>
5 not_pruned Other <conf_mat>
6 prune MexicanAmerican <conf_mat>
7 prune Other Hispanic <conf_mat>
8 prune Non-Hispanic White <conf_mat>
9 prune Non-Hispanic Black <conf_mat>
10 prune Other <conf_mat>str(cm_compare_groups,1)tibble [10 × 3] (S3: tbl_df/tbl/data.frame)cm_compare_groups[3,]$conf_mat[[1]] Truth
Prediction Diabetes No Diabetes
Diabetes 16 15
No Diabetes 14 215cm_compare_groups[3,c('model_type', 'ridreth1')]# A tibble: 1 × 2
model_type ridreth1
<chr> <fct>
1 not_pruned Non-Hispanic Whitesummary(cm_compare_groups$conf_mat[[3]])# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.888
2 kap binary 0.461
3 sens binary 0.533
4 spec binary 0.935
5 ppv binary 0.516
6 npv binary 0.939
7 mcc binary 0.462
8 j_index binary 0.468
9 bal_accuracy binary 0.734
10 detection_prevalence binary 0.119
11 precision binary 0.516
12 recall binary 0.533
13 f_meas binary 0.52522.10 Compare Groups Helper Function
Group_Model_Metic_Compare_helper_fun <- function(my_data, my_row_number, ...) {
group_var <- enquos(...)
row_of_data <- my_data %>%
filter(row_number() == my_row_number)
summary_stats <- summary(row_of_data$conf_mat[[1]]) %>%
mutate(join_key = 1)
row_of_data_2 <- row_of_data %>%
select(!!! group_var) %>%
mutate(join_key = 1)
output <- row_of_data_2 %>%
left_join(summary_stats, by = "join_key") %>%
select(-join_key)
return(output)
}22.10.1 Test Compare Groups Helper Function
Group_Model_Metic_Compare_helper_fun(cm_compare_groups, 3, model_type, ridreth1)# A tibble: 13 × 5
model_type ridreth1 .metric .estimator .estimate
<chr> <fct> <chr> <chr> <dbl>
1 not_pruned Non-Hispanic White accuracy binary 0.888
2 not_pruned Non-Hispanic White kap binary 0.461
3 not_pruned Non-Hispanic White sens binary 0.533
4 not_pruned Non-Hispanic White spec binary 0.935
5 not_pruned Non-Hispanic White ppv binary 0.516
6 not_pruned Non-Hispanic White npv binary 0.939
7 not_pruned Non-Hispanic White mcc binary 0.462
8 not_pruned Non-Hispanic White j_index binary 0.468
9 not_pruned Non-Hispanic White bal_accuracy binary 0.734
10 not_pruned Non-Hispanic White detection_prevalence binary 0.119
11 not_pruned Non-Hispanic White precision binary 0.516
12 not_pruned Non-Hispanic White recall binary 0.533
13 not_pruned Non-Hispanic White f_meas binary 0.52522.11 Apply Compare Groups Helper Function
list_to_apply_function <- 1:nrow(cm_compare_groups)
Final_Compare_Group_Table <- map_dfr(list_to_apply_function,
Group_Model_Metic_Compare_helper_fun,
my_data = cm_compare_groups,
model_type, ridreth1)
Final_Compare_Group_Table# A tibble: 130 × 5
model_type ridreth1 .metric .estimator .estimate
<chr> <fct> <chr> <chr> <dbl>
1 not_pruned MexicanAmerican accuracy binary 0.832
2 not_pruned MexicanAmerican kap binary 0.275
3 not_pruned MexicanAmerican sens binary 0.286
4 not_pruned MexicanAmerican spec binary 0.942
5 not_pruned MexicanAmerican ppv binary 0.5
6 not_pruned MexicanAmerican npv binary 0.867
7 not_pruned MexicanAmerican mcc binary 0.289
8 not_pruned MexicanAmerican j_index binary 0.228
9 not_pruned MexicanAmerican bal_accuracy binary 0.614
10 not_pruned MexicanAmerican detection_prevalence binary 0.096
# ℹ 120 more rows22.11.1 Bar Graph of Model Metrics by Race/Hispanic Origin, Model Type, and Diabetes
ggplot(Final_Compare_Group_Table, aes(.metric, .estimate, fill = model_type)) +
geom_bar(stat="identity", position=position_dodge()) +
coord_flip() +
facet_wrap(~ridreth1)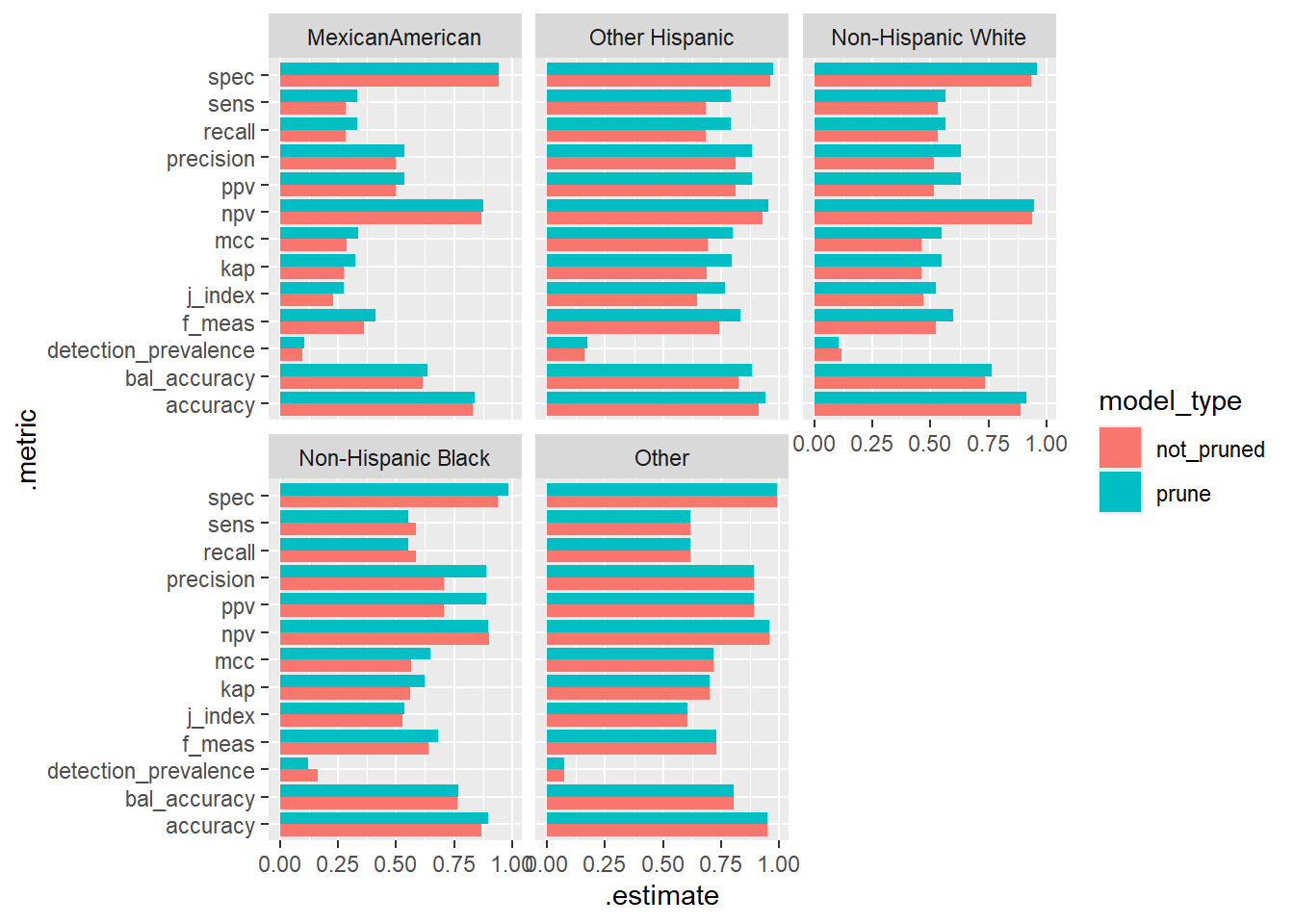
22.11.2 Multi-Group ROCs
Final_Compare_Group_Table.roc_auc <- diab_pop.test.stacked_dfs %>%
group_by(model_type,ridreth1) %>%
roc_auc(truth=diq010, pred_prob)
ggplot(Final_Compare_Group_Table.roc_auc, aes(ridreth1, .estimate, fill = model_type)) +
geom_bar(stat="identity", position=position_dodge()) +
coord_flip() 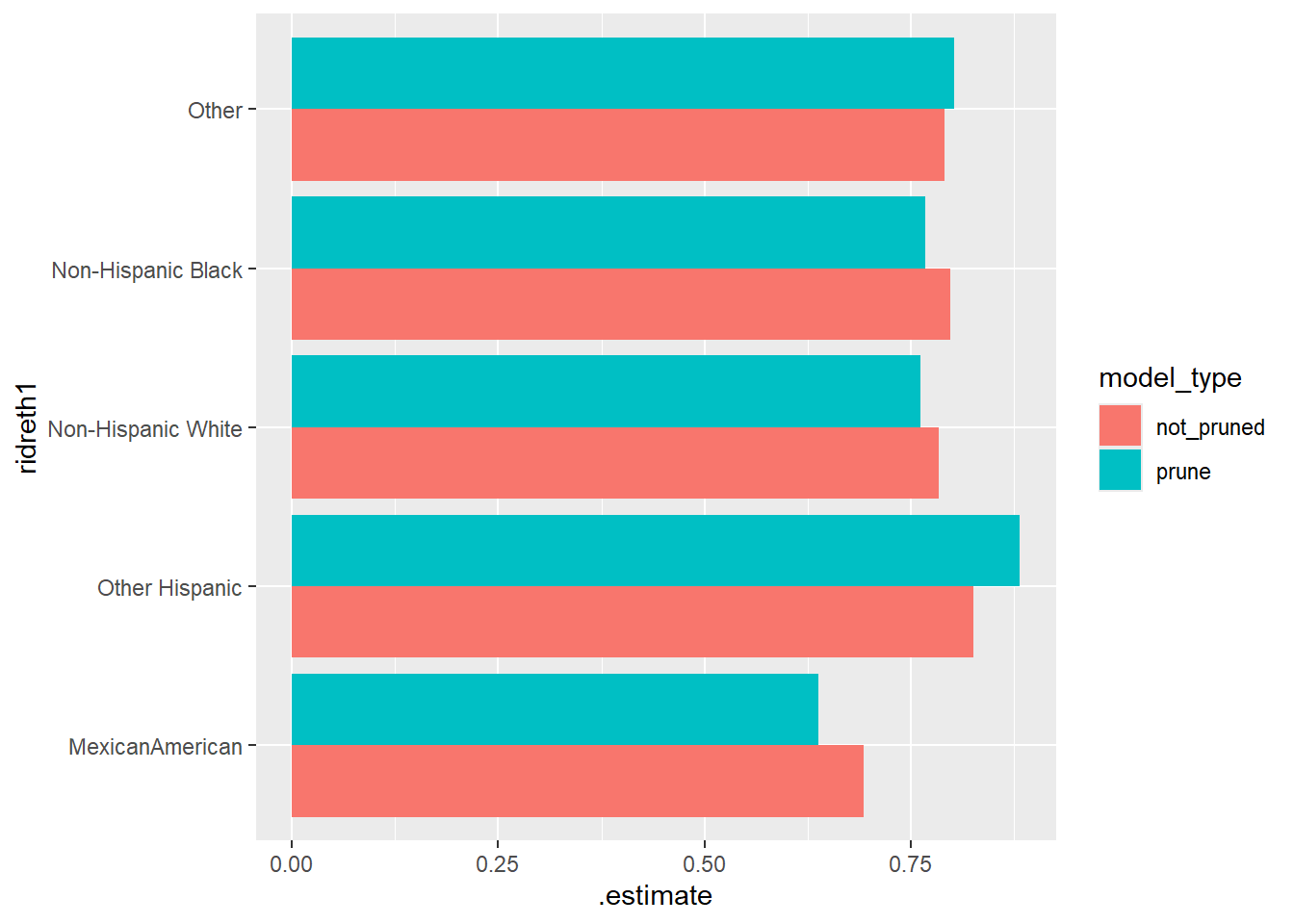
test_compare_groups_roc <- diab_pop.test.stacked_dfs %>%
group_by(model_type,ridreth1) %>%
roc_curve(truth=diq010, pred_prob)
autoplot(test_compare_groups_roc)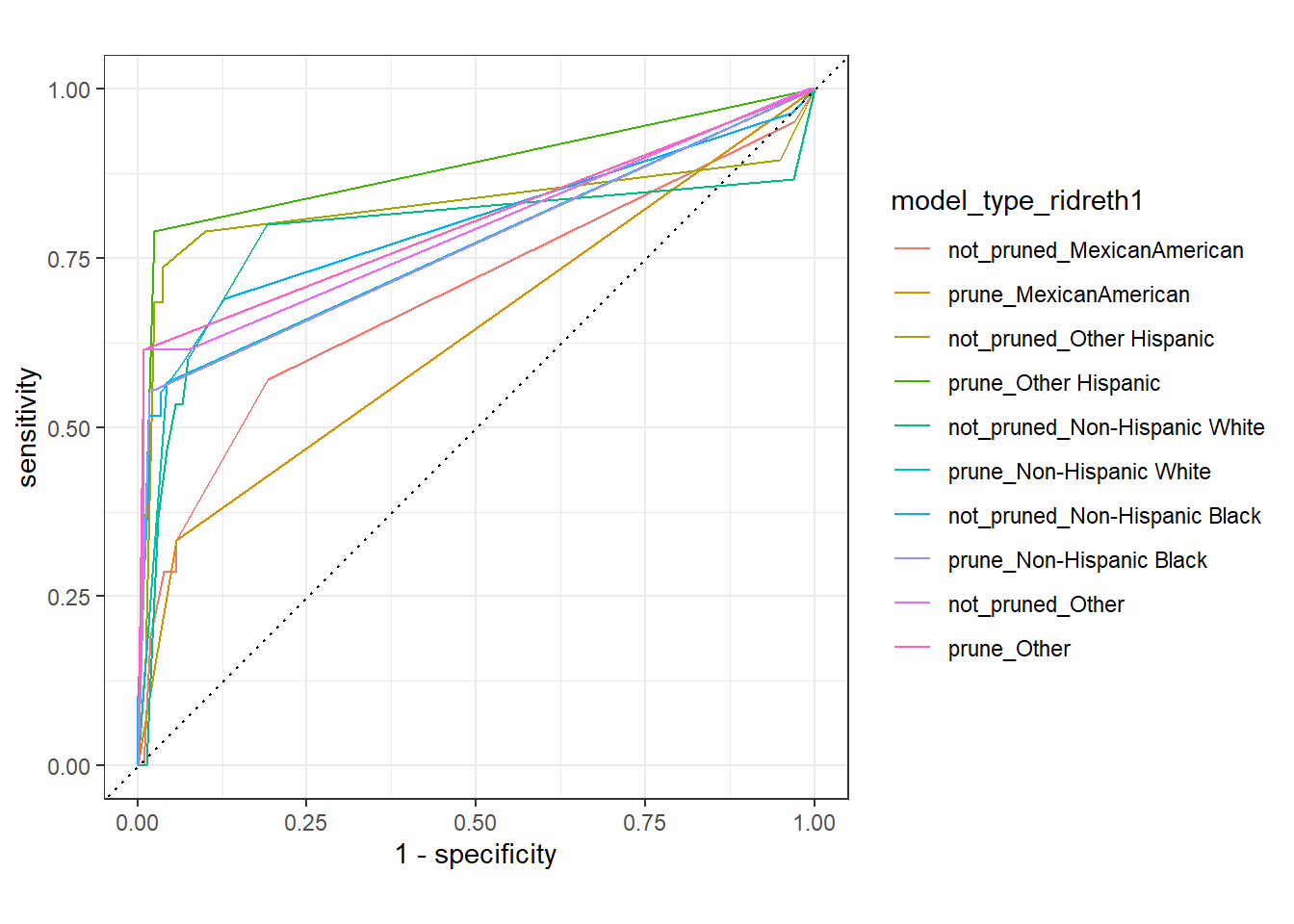
autoplot(test_compare_groups_roc) +
facet_wrap(~model_type)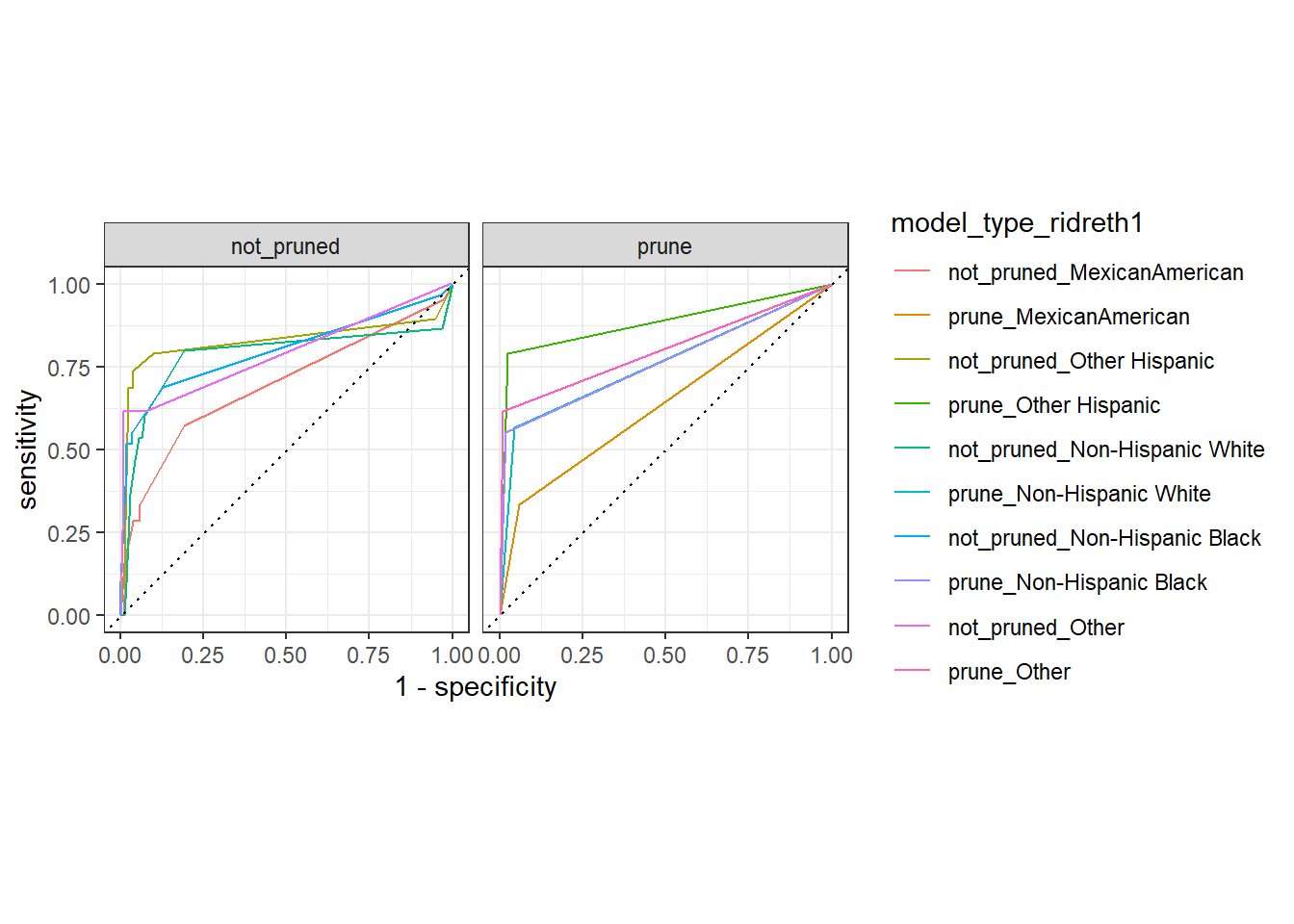
autoplot(test_compare_groups_roc) +
facet_wrap(~ridreth1)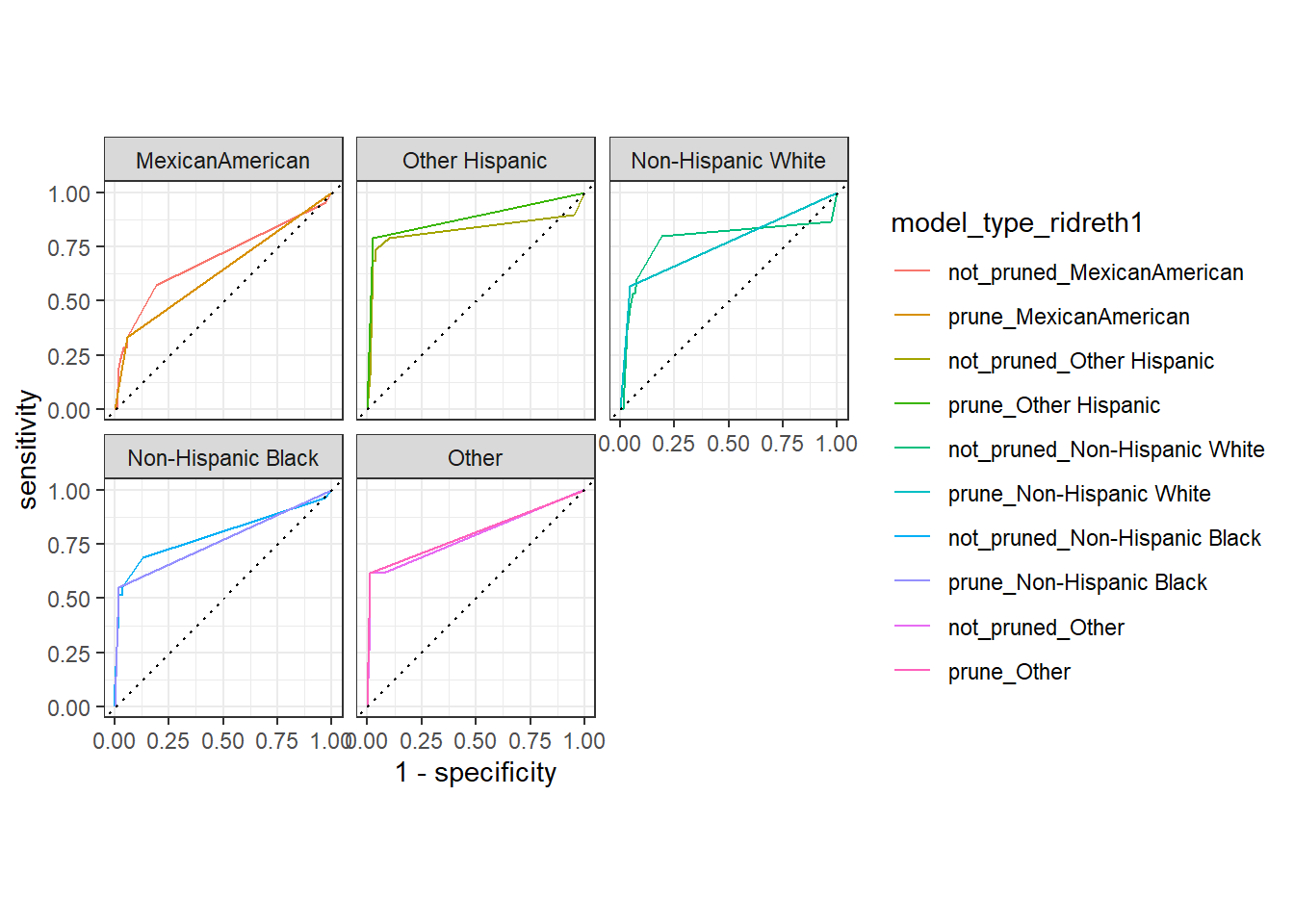
\(~\)
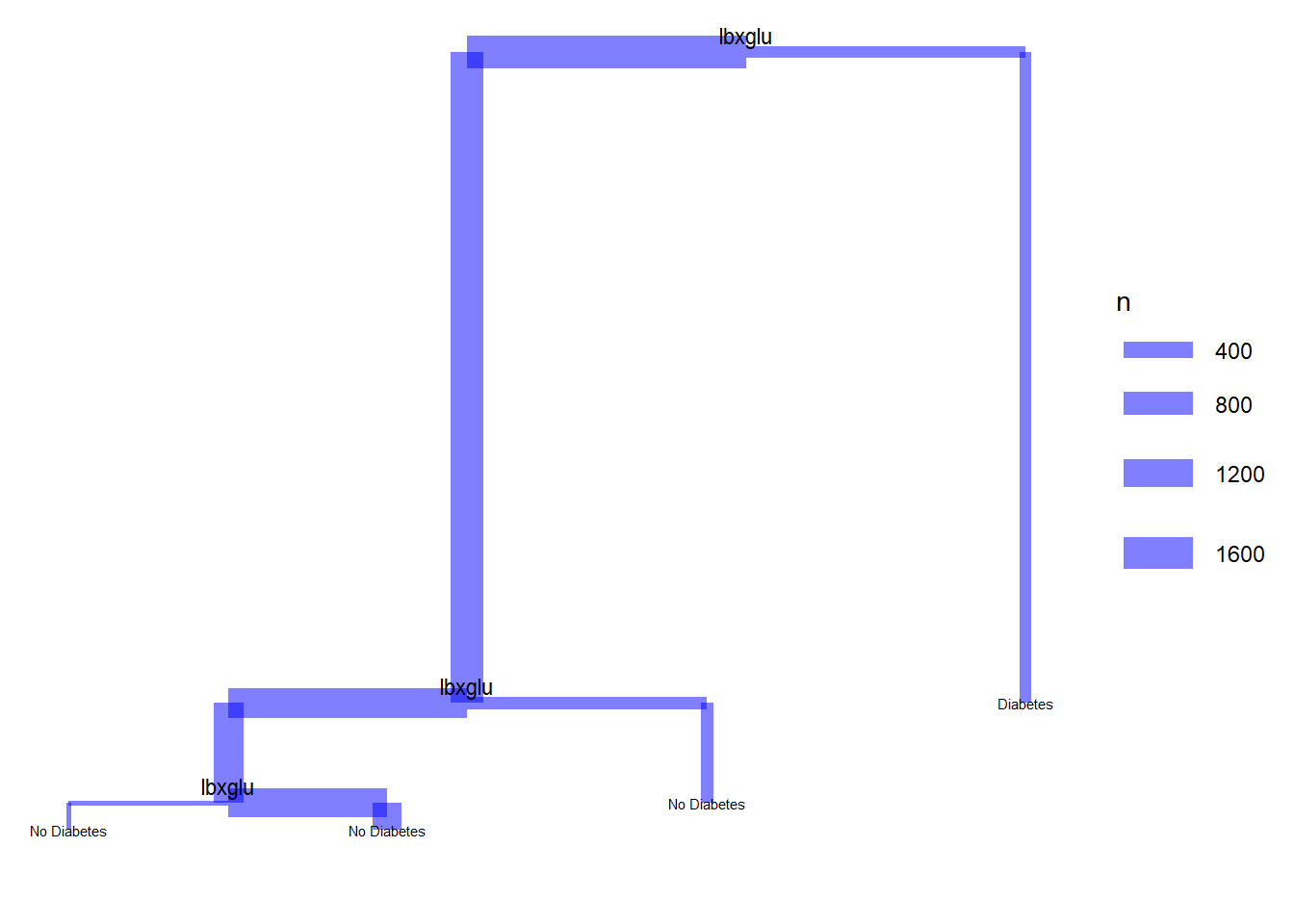
22.12 Dendrograms with ggdendro
\(~\)
library(ggplot2)
library(ggdendro)
library(tree)
model <- tree(diq010 ~ riagendr + ridreth1 + indhhin2 + dmdeduc2 + dmdmartl + bmxbmi + lbxglu,
data = diab_pop)
tree_data <- dendro_data(model)
segment(tree_data) %>%
ggplot() +
geom_segment(aes(x = x,
y = y,
xend = xend,
yend = yend,
size = n),
colour = "blue", alpha = 0.5) +
scale_size("n") +
geom_text(data = label(tree_data),
aes(x = x, y = y, label = label), vjust = -0.5, size = 3) +
geom_text(data = leaf_label(tree_data),
aes(x = x, y = y, label = label), vjust = 0.5, size = 2) +
theme_dendro()Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.