install_if_not <- function( list.of.packages ) {
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) { install.packages(new.packages) } else { print(paste0("the package '", list.of.packages , "' is already installed")) }
}
11 Logistic Regression with caret
11.1 Install if not Function
install_if_not("NHANES")[1] "the package 'NHANES' is already installed"install_if_not("RANN")[1] "the package 'RANN' is already installed"install_if_not("glmnet")[1] "the package 'glmnet' is already installed"11.2 NHANES Dataset
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors[1] 10000 75glimpse(data)Rows: 10,000
Columns: 75
$ ID <int> 51624, 51624, 51624, 51625, 51630, 51638, 51646, 5164…
$ SurveyYr <fct> 2009_10, 2009_10, 2009_10, 2009_10, 2009_10, 2009_10,…
$ Gender <fct> male, male, male, male, female, male, male, female, f…
$ Age <int> 34, 34, 34, 4, 49, 9, 8, 45, 45, 45, 66, 58, 54, 10, …
$ AgeDecade <fct> 30-39, 30-39, 30-39, 0-9, 40-49, 0-9, 0-9, 40…
$ AgeMonths <int> 409, 409, 409, 49, 596, 115, 101, 541, 541, 541, 795,…
$ Race1 <fct> White, White, White, Other, White, White, White, Whit…
$ Race3 <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ Education <fct> High School, High School, High School, NA, Some Colle…
$ MaritalStatus <fct> Married, Married, Married, NA, LivePartner, NA, NA, M…
$ HHIncome <fct> 25000-34999, 25000-34999, 25000-34999, 20000-24999, 3…
$ HHIncomeMid <int> 30000, 30000, 30000, 22500, 40000, 87500, 60000, 8750…
$ Poverty <dbl> 1.36, 1.36, 1.36, 1.07, 1.91, 1.84, 2.33, 5.00, 5.00,…
$ HomeRooms <int> 6, 6, 6, 9, 5, 6, 7, 6, 6, 6, 5, 10, 6, 10, 10, 4, 3,…
$ HomeOwn <fct> Own, Own, Own, Own, Rent, Rent, Own, Own, Own, Own, O…
$ Work <fct> NotWorking, NotWorking, NotWorking, NA, NotWorking, N…
$ Weight <dbl> 87.4, 87.4, 87.4, 17.0, 86.7, 29.8, 35.2, 75.7, 75.7,…
$ Length <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ HeadCirc <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ Height <dbl> 164.7, 164.7, 164.7, 105.4, 168.4, 133.1, 130.6, 166.…
$ BMI <dbl> 32.22, 32.22, 32.22, 15.30, 30.57, 16.82, 20.64, 27.2…
$ BMICatUnder20yrs <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ BMI_WHO <fct> 30.0_plus, 30.0_plus, 30.0_plus, 12.0_18.5, 30.0_plus…
$ Pulse <int> 70, 70, 70, NA, 86, 82, 72, 62, 62, 62, 60, 62, 76, 8…
$ BPSysAve <int> 113, 113, 113, NA, 112, 86, 107, 118, 118, 118, 111, …
$ BPDiaAve <int> 85, 85, 85, NA, 75, 47, 37, 64, 64, 64, 63, 74, 85, 6…
$ BPSys1 <int> 114, 114, 114, NA, 118, 84, 114, 106, 106, 106, 124, …
$ BPDia1 <int> 88, 88, 88, NA, 82, 50, 46, 62, 62, 62, 64, 76, 86, 6…
$ BPSys2 <int> 114, 114, 114, NA, 108, 84, 108, 118, 118, 118, 108, …
$ BPDia2 <int> 88, 88, 88, NA, 74, 50, 36, 68, 68, 68, 62, 72, 88, 6…
$ BPSys3 <int> 112, 112, 112, NA, 116, 88, 106, 118, 118, 118, 114, …
$ BPDia3 <int> 82, 82, 82, NA, 76, 44, 38, 60, 60, 60, 64, 76, 82, 7…
$ Testosterone <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ DirectChol <dbl> 1.29, 1.29, 1.29, NA, 1.16, 1.34, 1.55, 2.12, 2.12, 2…
$ TotChol <dbl> 3.49, 3.49, 3.49, NA, 6.70, 4.86, 4.09, 5.82, 5.82, 5…
$ UrineVol1 <int> 352, 352, 352, NA, 77, 123, 238, 106, 106, 106, 113, …
$ UrineFlow1 <dbl> NA, NA, NA, NA, 0.094, 1.538, 1.322, 1.116, 1.116, 1.…
$ UrineVol2 <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ UrineFlow2 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ Diabetes <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, N…
$ HealthGen <fct> Good, Good, Good, NA, Good, NA, NA, Vgood, Vgood, Vgo…
$ DaysPhysHlthBad <int> 0, 0, 0, NA, 0, NA, NA, 0, 0, 0, 10, 0, 4, NA, NA, 0,…
$ DaysMentHlthBad <int> 15, 15, 15, NA, 10, NA, NA, 3, 3, 3, 0, 0, 0, NA, NA,…
$ LittleInterest <fct> Most, Most, Most, NA, Several, NA, NA, None, None, No…
$ Depressed <fct> Several, Several, Several, NA, Several, NA, NA, None,…
$ nPregnancies <int> NA, NA, NA, NA, 2, NA, NA, 1, 1, 1, NA, NA, NA, NA, N…
$ nBabies <int> NA, NA, NA, NA, 2, NA, NA, NA, NA, NA, NA, NA, NA, NA…
$ Age1stBaby <int> NA, NA, NA, NA, 27, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ SleepHrsNight <int> 4, 4, 4, NA, 8, NA, NA, 8, 8, 8, 7, 5, 4, NA, 5, 7, N…
$ SleepTrouble <fct> Yes, Yes, Yes, NA, Yes, NA, NA, No, No, No, No, No, Y…
$ PhysActive <fct> No, No, No, NA, No, NA, NA, Yes, Yes, Yes, Yes, Yes, …
$ PhysActiveDays <int> NA, NA, NA, NA, NA, NA, NA, 5, 5, 5, 7, 5, 1, NA, 2, …
$ TVHrsDay <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ CompHrsDay <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ TVHrsDayChild <int> NA, NA, NA, 4, NA, 5, 1, NA, NA, NA, NA, NA, NA, 4, N…
$ CompHrsDayChild <int> NA, NA, NA, 1, NA, 0, 6, NA, NA, NA, NA, NA, NA, 3, N…
$ Alcohol12PlusYr <fct> Yes, Yes, Yes, NA, Yes, NA, NA, Yes, Yes, Yes, Yes, Y…
$ AlcoholDay <int> NA, NA, NA, NA, 2, NA, NA, 3, 3, 3, 1, 2, 6, NA, NA, …
$ AlcoholYear <int> 0, 0, 0, NA, 20, NA, NA, 52, 52, 52, 100, 104, 364, N…
$ SmokeNow <fct> No, No, No, NA, Yes, NA, NA, NA, NA, NA, No, NA, NA, …
$ Smoke100 <fct> Yes, Yes, Yes, NA, Yes, NA, NA, No, No, No, Yes, No, …
$ Smoke100n <fct> Smoker, Smoker, Smoker, NA, Smoker, NA, NA, Non-Smoke…
$ SmokeAge <int> 18, 18, 18, NA, 38, NA, NA, NA, NA, NA, 13, NA, NA, N…
$ Marijuana <fct> Yes, Yes, Yes, NA, Yes, NA, NA, Yes, Yes, Yes, NA, Ye…
$ AgeFirstMarij <int> 17, 17, 17, NA, 18, NA, NA, 13, 13, 13, NA, 19, 15, N…
$ RegularMarij <fct> No, No, No, NA, No, NA, NA, No, No, No, NA, Yes, Yes,…
$ AgeRegMarij <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 20, 15, N…
$ HardDrugs <fct> Yes, Yes, Yes, NA, Yes, NA, NA, No, No, No, No, Yes, …
$ SexEver <fct> Yes, Yes, Yes, NA, Yes, NA, NA, Yes, Yes, Yes, Yes, Y…
$ SexAge <int> 16, 16, 16, NA, 12, NA, NA, 13, 13, 13, 17, 22, 12, N…
$ SexNumPartnLife <int> 8, 8, 8, NA, 10, NA, NA, 20, 20, 20, 15, 7, 100, NA, …
$ SexNumPartYear <int> 1, 1, 1, NA, 1, NA, NA, 0, 0, 0, NA, 1, 1, NA, NA, 1,…
$ SameSex <fct> No, No, No, NA, Yes, NA, NA, Yes, Yes, Yes, No, No, N…
$ SexOrientation <fct> Heterosexual, Heterosexual, Heterosexual, NA, Heteros…
$ PregnantNow <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…11.3 Missing data
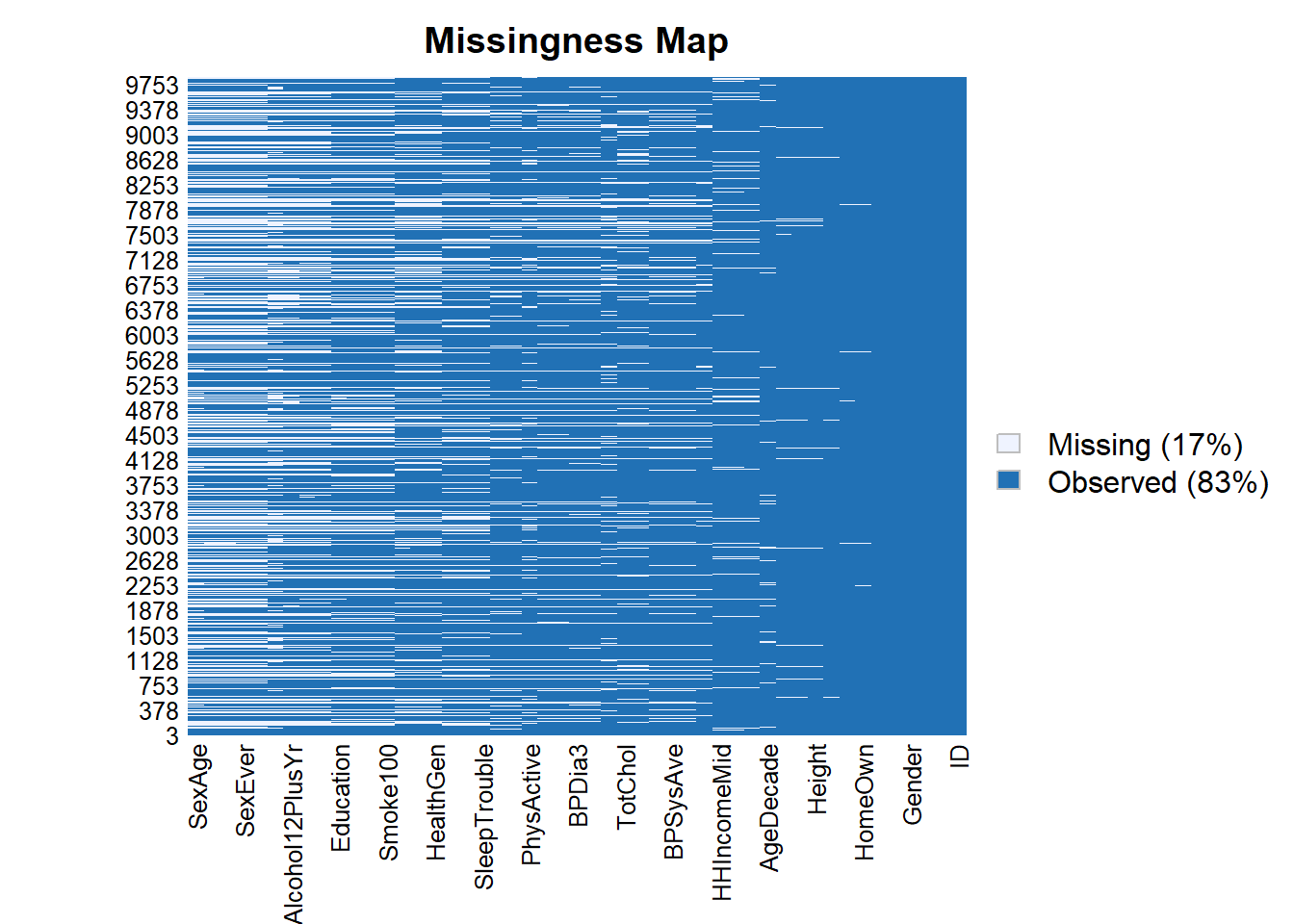
Amelia::missmap(data)Warning: Unknown or uninitialised column: `arguments`.
Unknown or uninitialised column: `arguments`.Warning: Unknown or uninitialised column: `imputations`.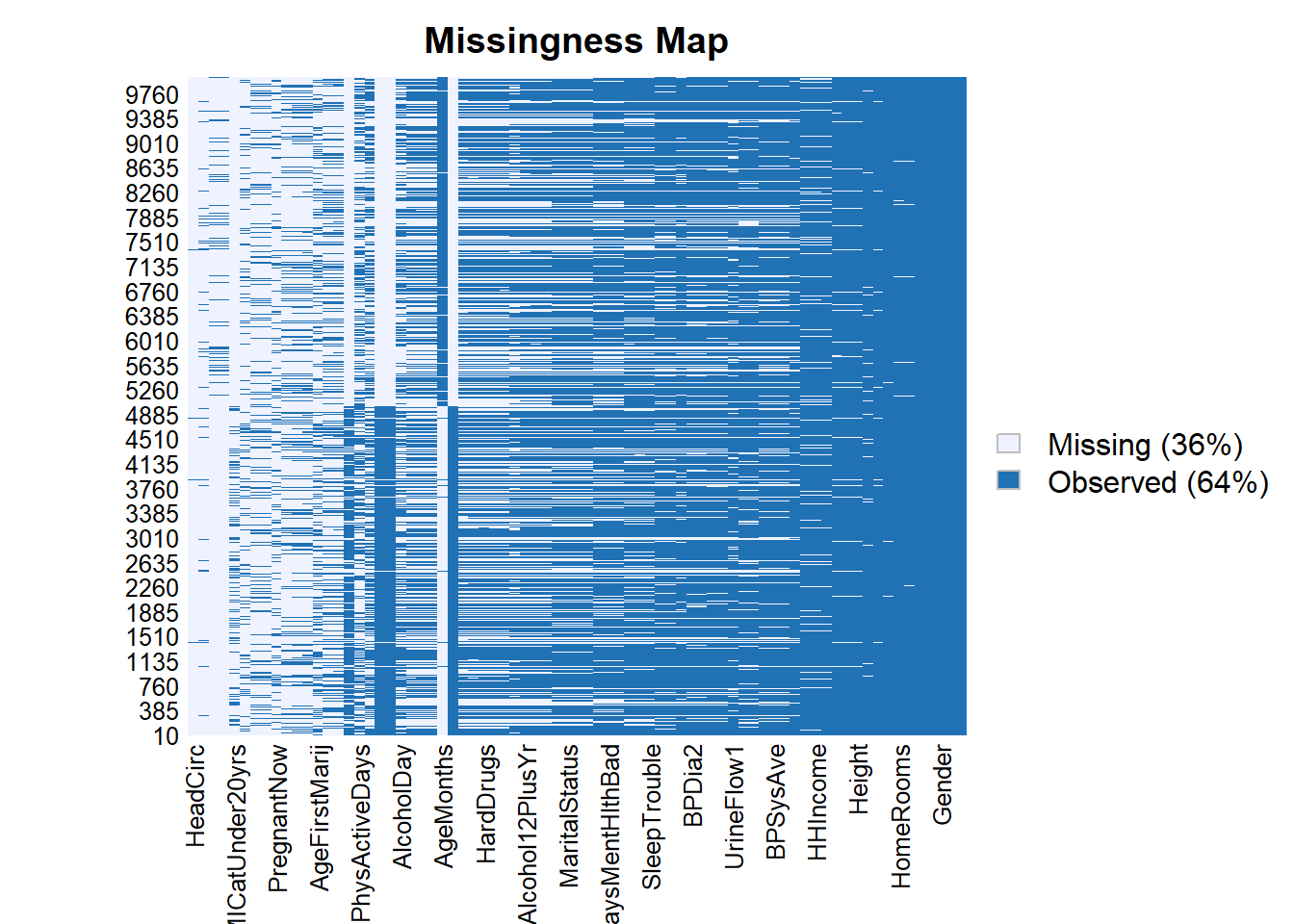
SumNa <- function(col){sum(is.na(col))}
data.sum <- data %>%
summarise_all(SumNa) %>%
tidyr::gather(key='feature', value='SumNa') %>%
arrange(-SumNa) %>%
mutate(PctNa = SumNa/nrow(data))
data.sum# A tibble: 75 × 3
feature SumNa PctNa
<chr> <int> <dbl>
1 HeadCirc 9912 0.991
2 Length 9457 0.946
3 TVHrsDayChild 9347 0.935
4 CompHrsDayChild 9347 0.935
5 BMICatUnder20yrs 8726 0.873
6 AgeRegMarij 8634 0.863
7 UrineFlow2 8524 0.852
8 UrineVol2 8522 0.852
9 PregnantNow 8304 0.830
10 Age1stBaby 8116 0.812
# ℹ 65 more rowsdata.sum2 <- data.sum %>%
filter(! (feature %in% c('ID','Diabetes'))) %>%
filter(PctNa < .45)
data.sum2$feature [1] "SexAge" "SexNumPartnLife" "HardDrugs" "SexEver"
[5] "SameSex" "AlcoholYear" "Alcohol12PlusYr" "LittleInterest"
[9] "Depressed" "Education" "MaritalStatus" "Smoke100"
[13] "Smoke100n" "DaysPhysHlthBad" "DaysMentHlthBad" "HealthGen"
[17] "SleepHrsNight" "Work" "SleepTrouble" "BPSys1"
[ reached getOption("max.print") -- omitted 27 entries ]11.4 Split Data
11.5 caret
11.6 dummyVars
dV <- dummyVars(Diabetes ~ ., TRAIN)
dVDummy Variable Object
Formula: Diabetes ~ .
49 variables, 22 factors
Variables and levels will be separated by '.'
A less than full rank encoding is useddV.TRAIN <- predict(dV, TRAIN)Warning in model.frame.default(Terms, newdata, na.action = na.action, xlev =
object$lvls): variable 'Diabetes' is not a factor num [1:7393, 1:104] 53639 54356 54171 64860 66251 ...
- attr(*, "dimnames")=List of 2
..$ : chr [1:7393] "1" "2" "3" "4" ...
..$ : chr [1:104] "ID" "SexAge" "SexNumPartnLife" "HardDrugs.No" ...11.7 impute, center, & scale
pP <- preProcess(dV.TRAIN,
method = c('knnImpute', 'center', 'scale'))
pPCreated from 2947 samples and 104 variables
Pre-processing:
- centered (104)
- ignored (0)
- 5 nearest neighbor imputation (104)
- scaled (104)pP.dV.TRAIN <- predict(pP, dV.TRAIN)11.8 train model
trControl <- trainControl(method = 'repeatedcv',
number = 10,
repeats = 5,
search = 'random')
logit.CV <- train(x= pP.dV.TRAIN , y= TRAIN$Diabetes,
method = 'glm',
trControl = trControl,
family = 'binomial' )Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurredlogit.CVGeneralized Linear Model
7393 samples
104 predictor
2 classes: 'No', 'Yes'
No pre-processing
Resampling: Cross-Validated (10 fold, repeated 5 times)
Summary of sample sizes: 6654, 6653, 6654, 6653, 6654, 6653, ...
Resampling results:
Accuracy Kappa
0.9273642 0.238551611.9 score test & train data
11.9.1 score test
# score TEST
dV.TEST <- predict(dV, TEST)Warning in model.frame.default(Terms, newdata, na.action = na.action, xlev =
object$lvls): variable 'Diabetes' is not a factor11.9.2 score train
11.10 yardstick
Attaching package: 'yardstick'The following objects are masked from 'package:caret':
precision, recall, sensitivity, specificityThe following object is masked from 'package:readr':
specTRAIN_TEST_scored_auc <- TRAIN_TEST_scored %>%
group_by(data) %>%
roc_auc(truth=Diabetes,Yes, event_level = 'second')
TRAIN_TEST_scored %>%
left_join(TRAIN_TEST_scored_auc) %>%
mutate(data = paste(data, " AUC: ", round(.estimate,3))) %>%
group_by(data) %>%
roc_curve(truth=Diabetes,Yes, event_level = 'second') %>%
autoplot()Joining with `by = join_by(data)`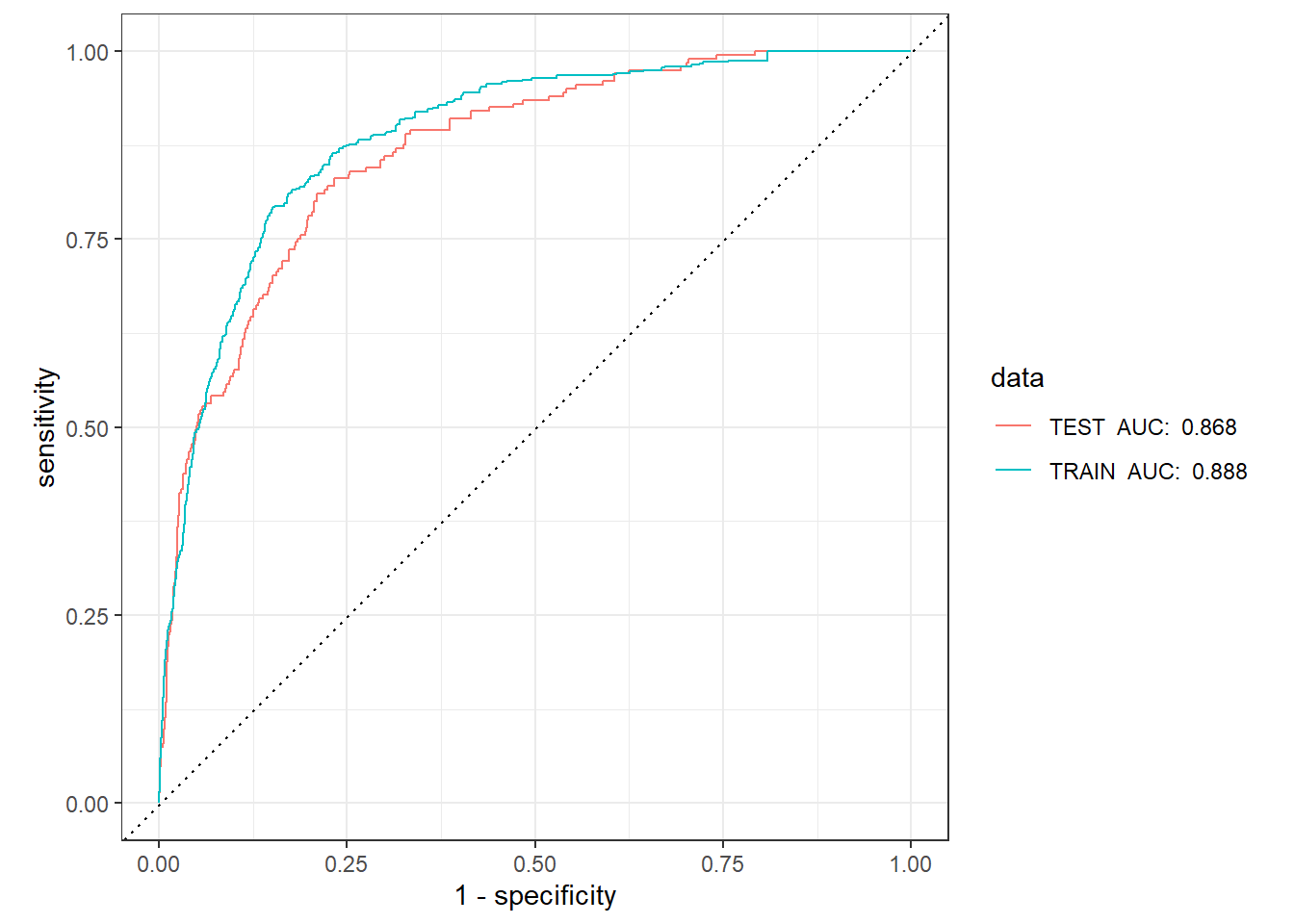
conf_mat.TRAIN_TEST_scored <- TRAIN_TEST_scored %>%
group_by(data) %>%
conf_mat(truth=Diabetes,class)
conf_mat.TRAIN_TEST_scored# A tibble: 2 × 2
data conf_mat
<chr> <list>
1 TEST <conf_mat>
2 TRAIN <conf_mat>conf_mat.TRAIN_TEST_scored$conf_mat[[1]]
Truth
Prediction No Yes
No 2240 167
Yes 24 34
[[2]]
Truth
Prediction No Yes
No 6776 452
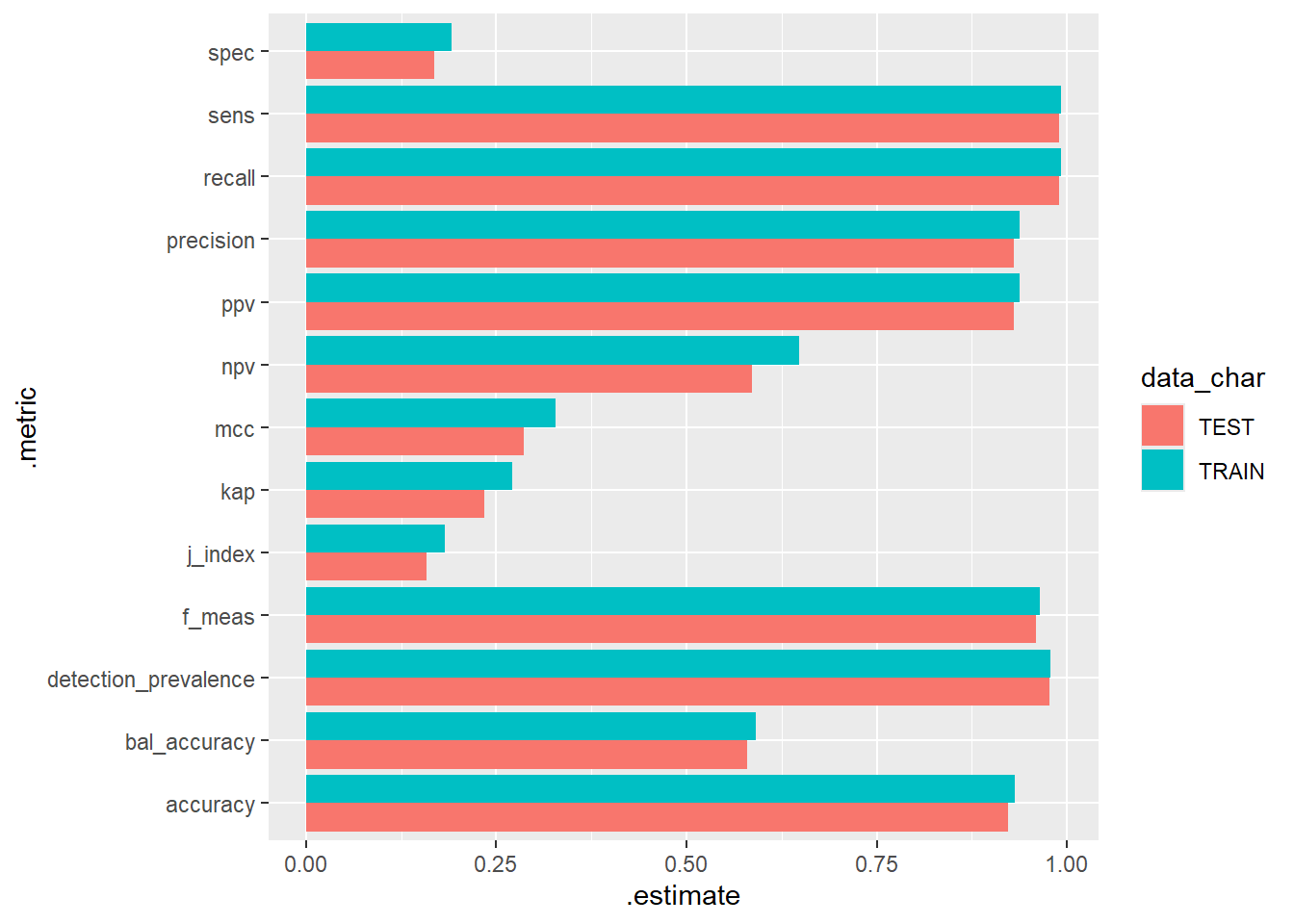
Yes 58 107metrics.TRAIN_TEST_scored <- map_dfr(conf_mat.TRAIN_TEST_scored$conf_mat,
summary,
.id="data") %>%
mutate(data_char = case_when(data == 1 ~ "TEST",
data == 2 ~ "TRAIN") )
library('ggplot2')
metrics.TRAIN_TEST_scored %>%
ggplot(aes(x = .metric, y=.estimate, fill= data_char)) +
geom_bar(stat="identity", position=position_dodge()) +
coord_flip()