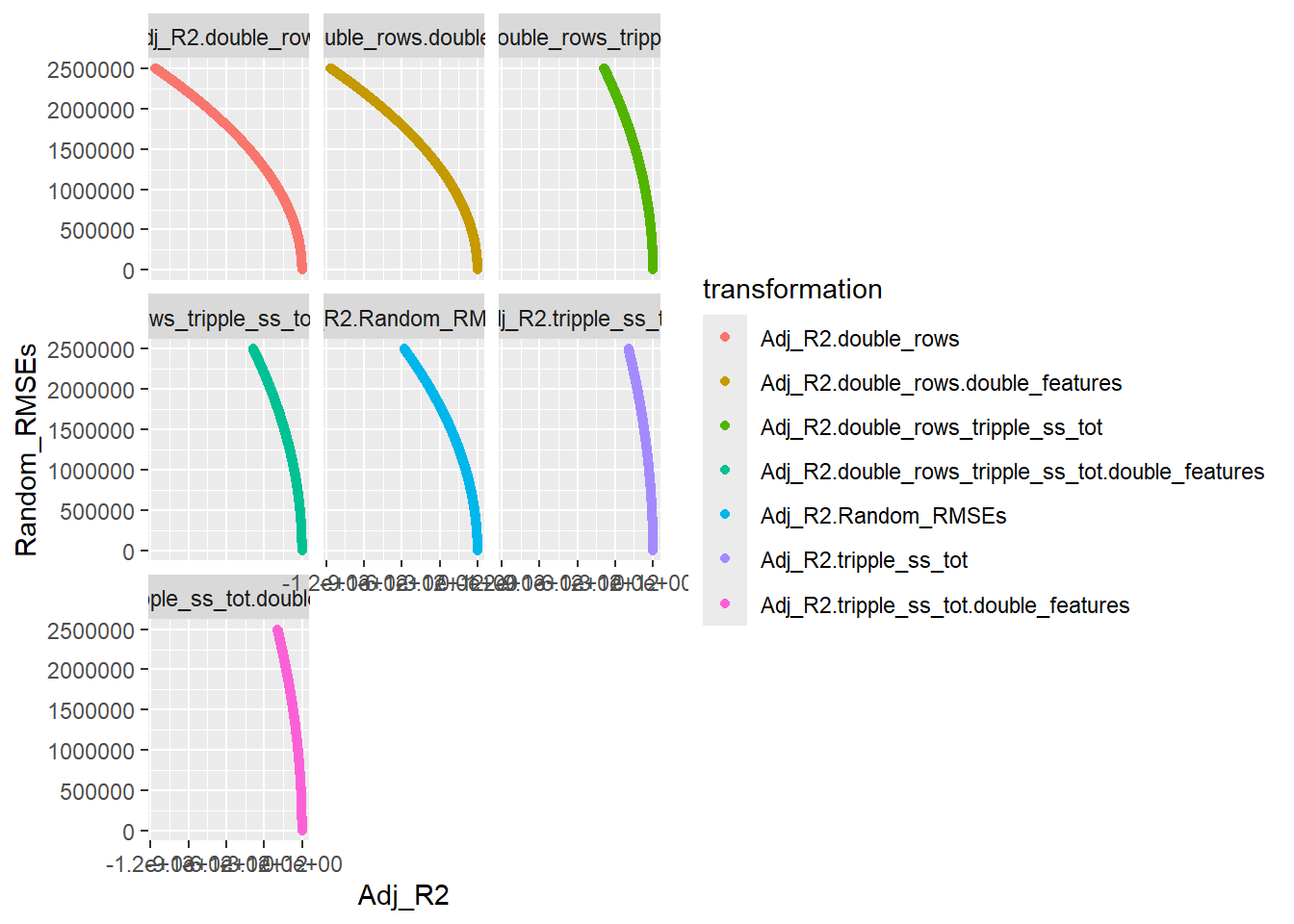
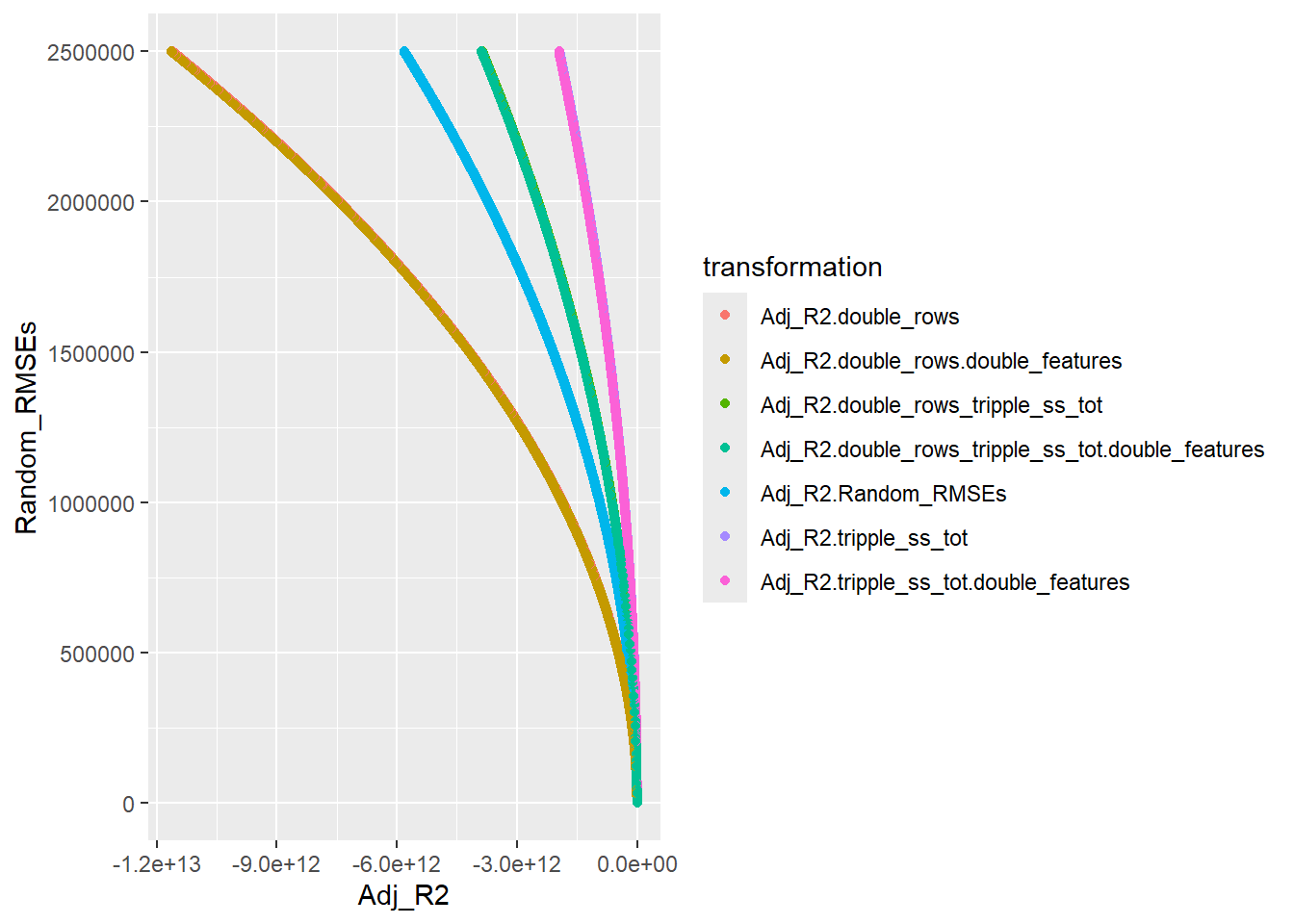
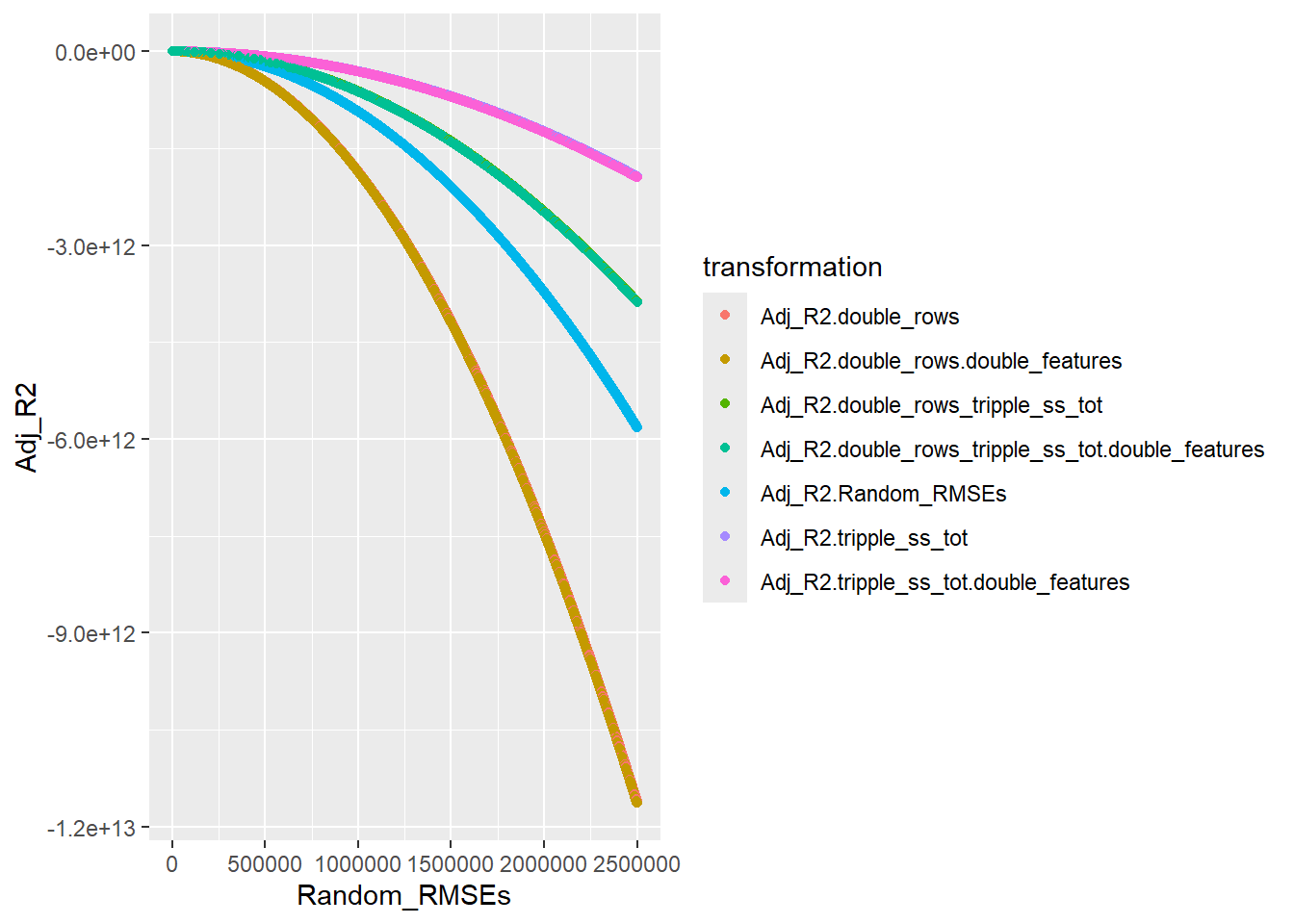
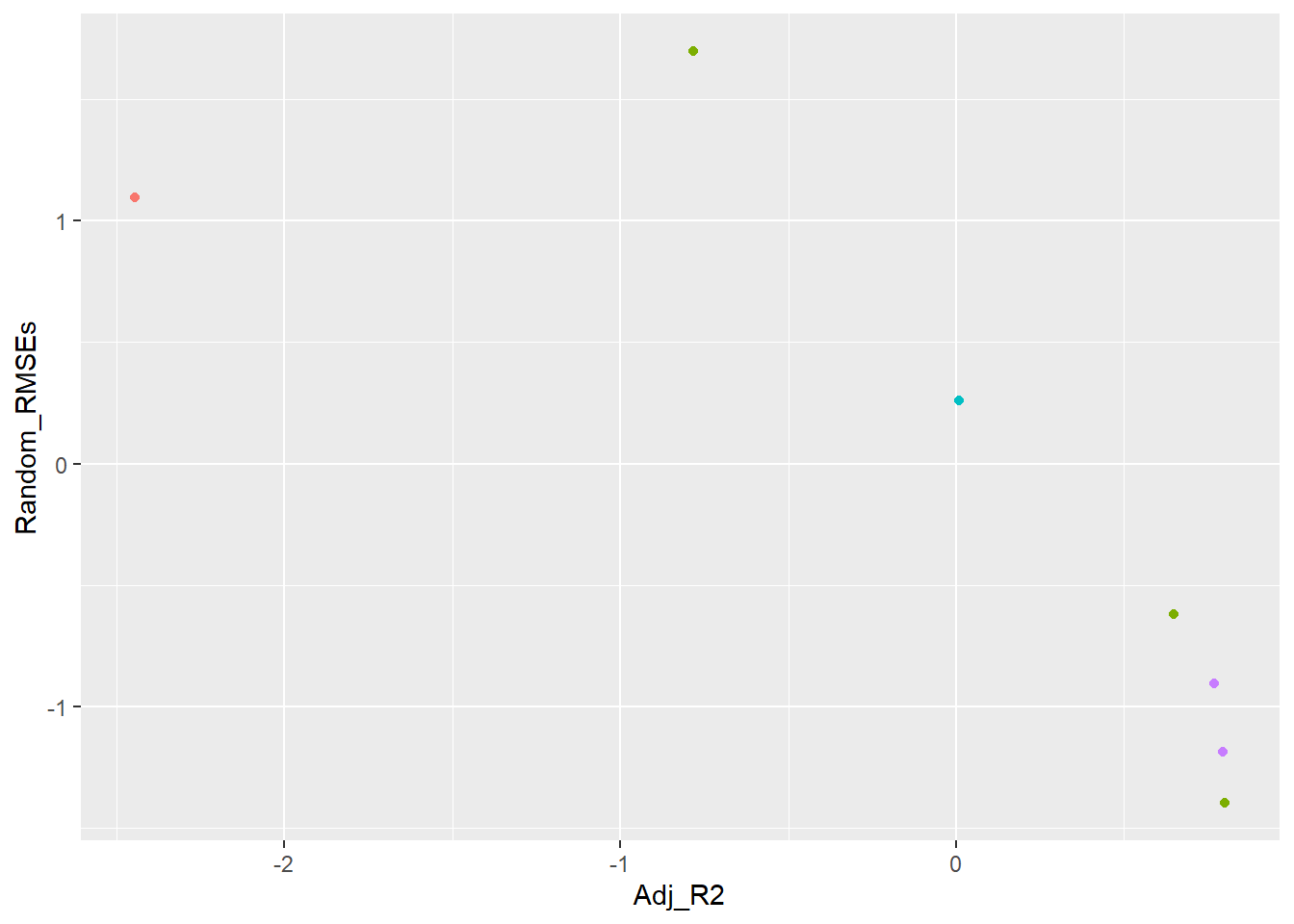
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsLoading required package: lattice
Attaching package: 'caret'
The following object is masked from 'package:purrr':
liftdiab_pop <- readRDS('C:/Users/jkyle/Documents/GitHub/Intro_Jeff_Data_Science/DATA/diab_pop.RDS') %>%
select(-seqn)
glimpse(diab_pop)Rows: 5,719
Columns: 9
$ riagendr <fct> Male, Male, Male, Female, Female, Female, Male, Female, Male,…
$ ridageyr <dbl> 62, 53, 78, 56, 42, 72, 22, 32, 56, 46, 45, 30, 67, 67, 57, 8…
$ ridreth1 <fct> Non-Hispanic White, Non-Hispanic White, Non-Hispanic White, N…
$ dmdeduc2 <fct> College grad or above, High school graduate/GED, High school …
$ dmdmartl <fct> Married, Divorced, Married, Living with partner, Divorced, Se…
$ indhhin2 <fct> "$65,000-$74,999", "$15,000-$19,999", "$20,000-$24,999", "$65…
$ bmxbmi <dbl> 27.8, 30.8, 28.8, 42.4, 20.3, 28.6, 28.0, 28.2, 33.6, 27.6, 2…
$ diq010 <fct> Diabetes, No Diabetes, Diabetes, No Diabetes, No Diabetes, No…
$ lbxglu <dbl> NA, 101, 84, NA, 84, 107, 95, NA, NA, NA, 84, NA, 130, 284, 3…