6 Naïve Bayes
6.0.1 Reminders about the Data
tibble::tribble(
~"Variable in Data", ~"Definition", ~"Data Type",
'seqn', 'Respondent sequence number', 'Identifier',
'riagendr', 'Gender', 'Categorical',
'ridageyr', 'Age in years at screening', 'Continuous / Numerical',
'ridreth1', 'Race/Hispanic origin', 'Categorical',
'dmdeduc2', 'Education level', 'Adults 20+ - Categorical',
'dmdmartl', 'Marital status', 'Categorical',
'indhhin2', 'Annual household income', 'Categorical',
'bmxbmi', 'Body Mass Index (kg/m**2)', 'Continuous / Numerical',
'diq010', 'Doctor diagnosed diabetes', 'Categorical',
'lbxglu', 'Fasting Glucose (mg/dL)', 'Continuous / Numerical'
) |>
knitr::kable()| Variable in Data | Definition | Data Type |
|---|---|---|
| seqn | Respondent sequence number | Identifier |
| riagendr | Gender | Categorical |
| ridageyr | Age in years at screening | Continuous / Numerical |
| ridreth1 | Race/Hispanic origin | Categorical |
| dmdeduc2 | Education level | Adults 20+ - Categorical |
| dmdmartl | Marital status | Categorical |
| indhhin2 | Annual household income | Categorical |
| bmxbmi | Body Mass Index (kg/m**2) | Continuous / Numerical |
| diq010 | Doctor diagnosed diabetes | Categorical |
| lbxglu | Fasting Glucose (mg/dL) | Continuous / Numerical |
6.0.2 Install if not Function
install_if_not <- function( list.of.packages ) {
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) { install.packages(new.packages) } else { print(paste0("the package '", list.of.packages , "' is already installed")) }
}
6.1 The e1071 package
6.2 Split Data
Loading required package: ggplot2Loading required package: latticediab_pop.no_na <- na.omit(diab_pop)
trainIndex <- createDataPartition(diab_pop.no_na$diq010,
list = FALSE ,
p = .8)
train <- diab_pop.no_na[trainIndex, ]
test <- diab_pop.no_na[-trainIndex, ]6.3 Make Formula
6.4 Train Model
model_nb <- naiveBayes( as.formula(my_formula) , data=train)
summary(model_nb) Length Class Mode
apriori 2 table numeric
tables 8 -none- list
levels 2 -none- character
isnumeric 8 -none- logical
call 4 -none- call 6.5 Score
6.6 yardstick
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, union
Attaching package: 'yardstick'The following objects are masked from 'package:caret':
precision, recall, sensitivity, specificity Truth
Prediction Diabetes No Diabetes
Diabetes 25 4
No Diabetes 31 315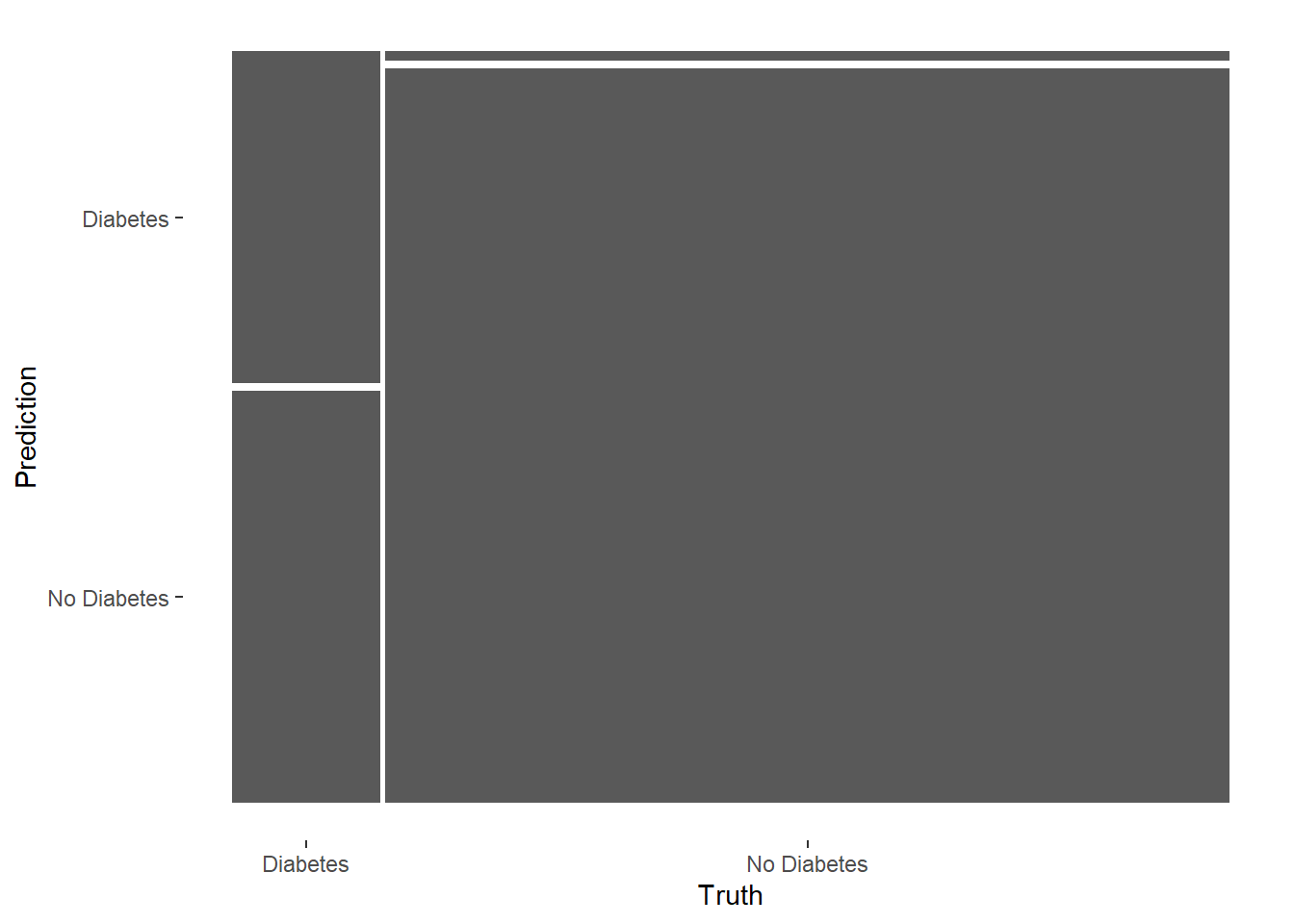
# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy binary 0.907
2 kap binary 0.542
3 sens binary 0.446
4 spec binary 0.987
5 ppv binary 0.862
6 npv binary 0.910
7 mcc binary 0.579
8 j_index binary 0.434
9 bal_accuracy binary 0.717
10 detection_prevalence binary 0.0773
11 precision binary 0.862
12 recall binary 0.446
13 f_meas binary 0.588 test.scored %>%
conf_mat(truth=diq010, class) %>%
summary() %>%
ggplot(aes(y=.metric, x=.estimate, fill=.metric)) +
geom_bar(stat="identity")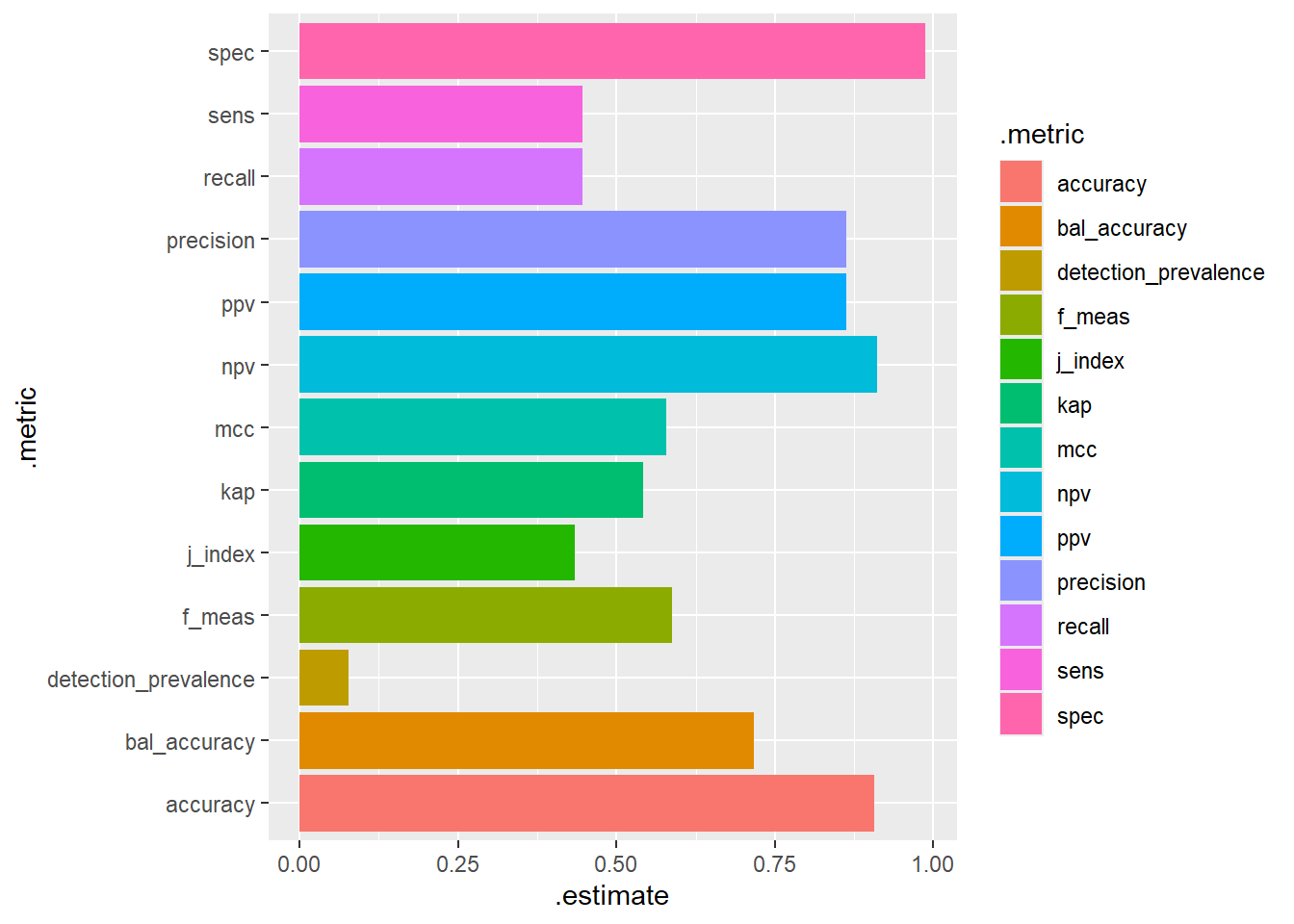
test.scored %>%
roc_curve(truth=diq010, Diabetes) %>%
mutate(FPR = 1 - specificity) %>%
mutate(TPR = sensitivity) %>%
ggplot(aes(x=FPR, y=TPR)) +
geom_line() +
annotate("text",
x=.94, y=0,
label= paste("AUC: ",round(roc_auc(test.scored, truth=diq010, Diabetes)$.estimate,3))) +
ggtitle("ROC Curve")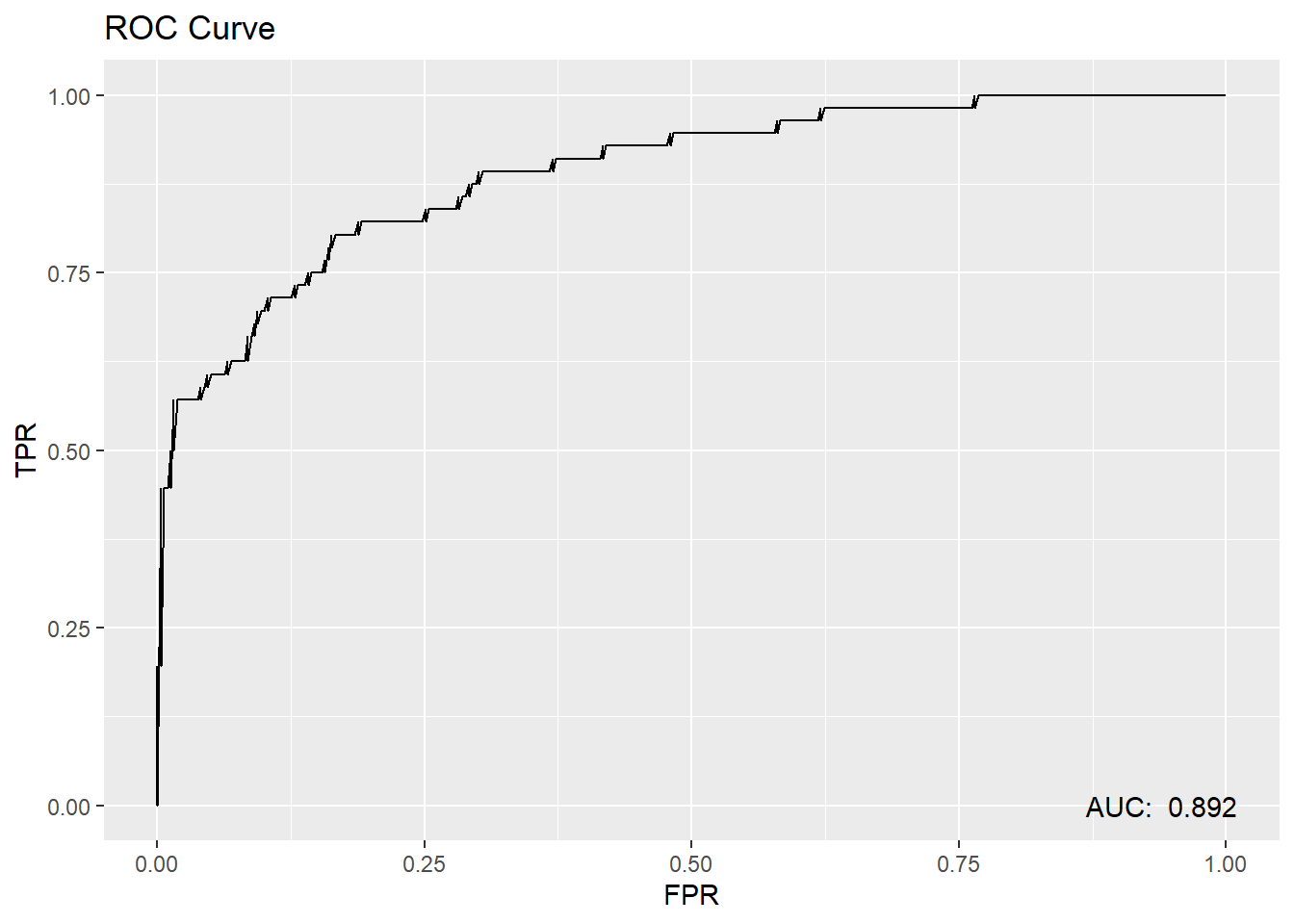
test.scored %>%
pr_curve(truth=diq010, Diabetes) %>%
ggplot(aes(x=recall, y=precision)) +
geom_line() +
annotate("text",
x=.94, y=0,
label= paste("PR_AUC: ",round(pr_auc(test.scored, truth=diq010, Diabetes)$.estimate,3))) +
ggtitle("PR Curve")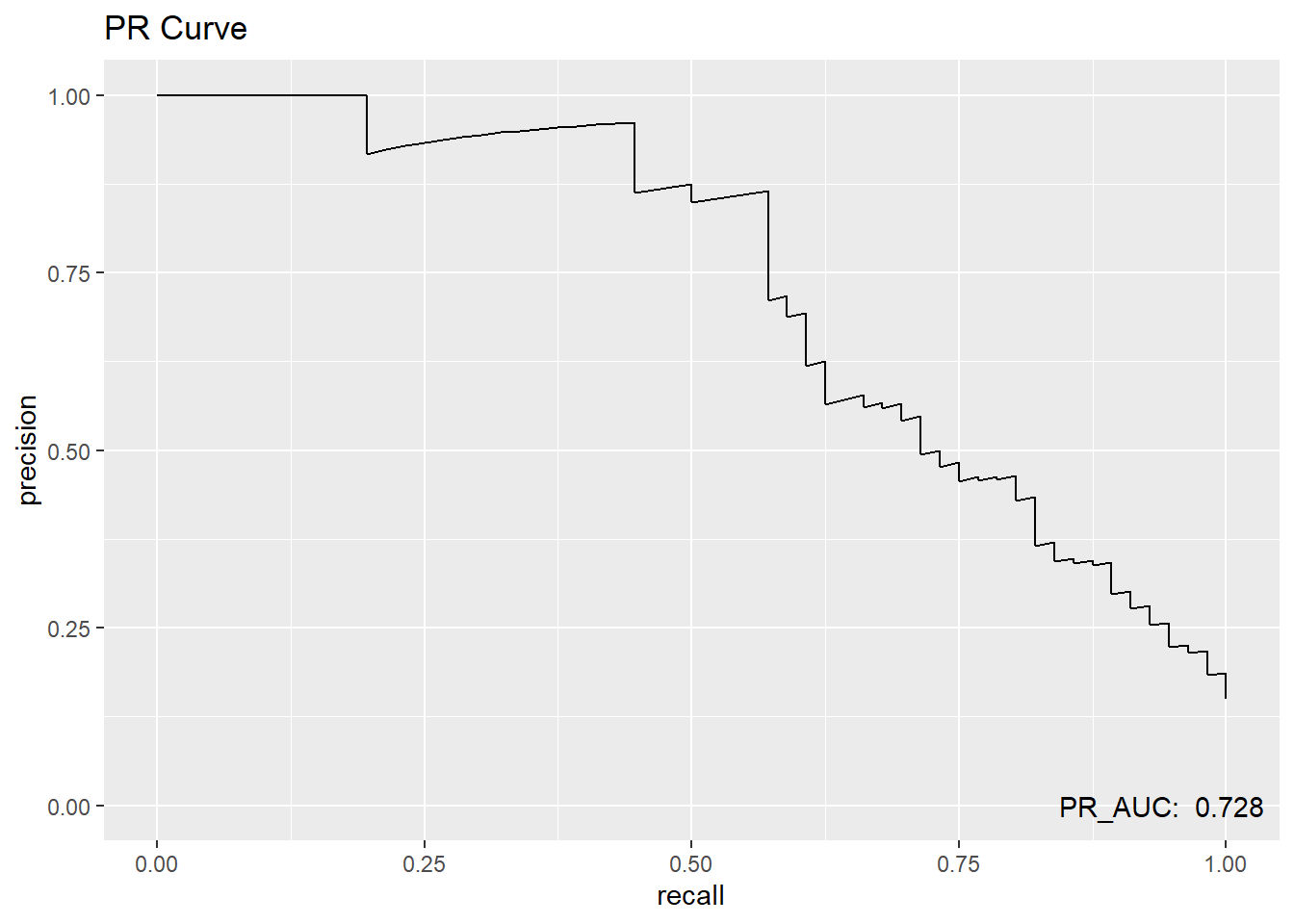
test.scored %>%
gain_curve(truth=diq010, Diabetes) %>%
autoplot()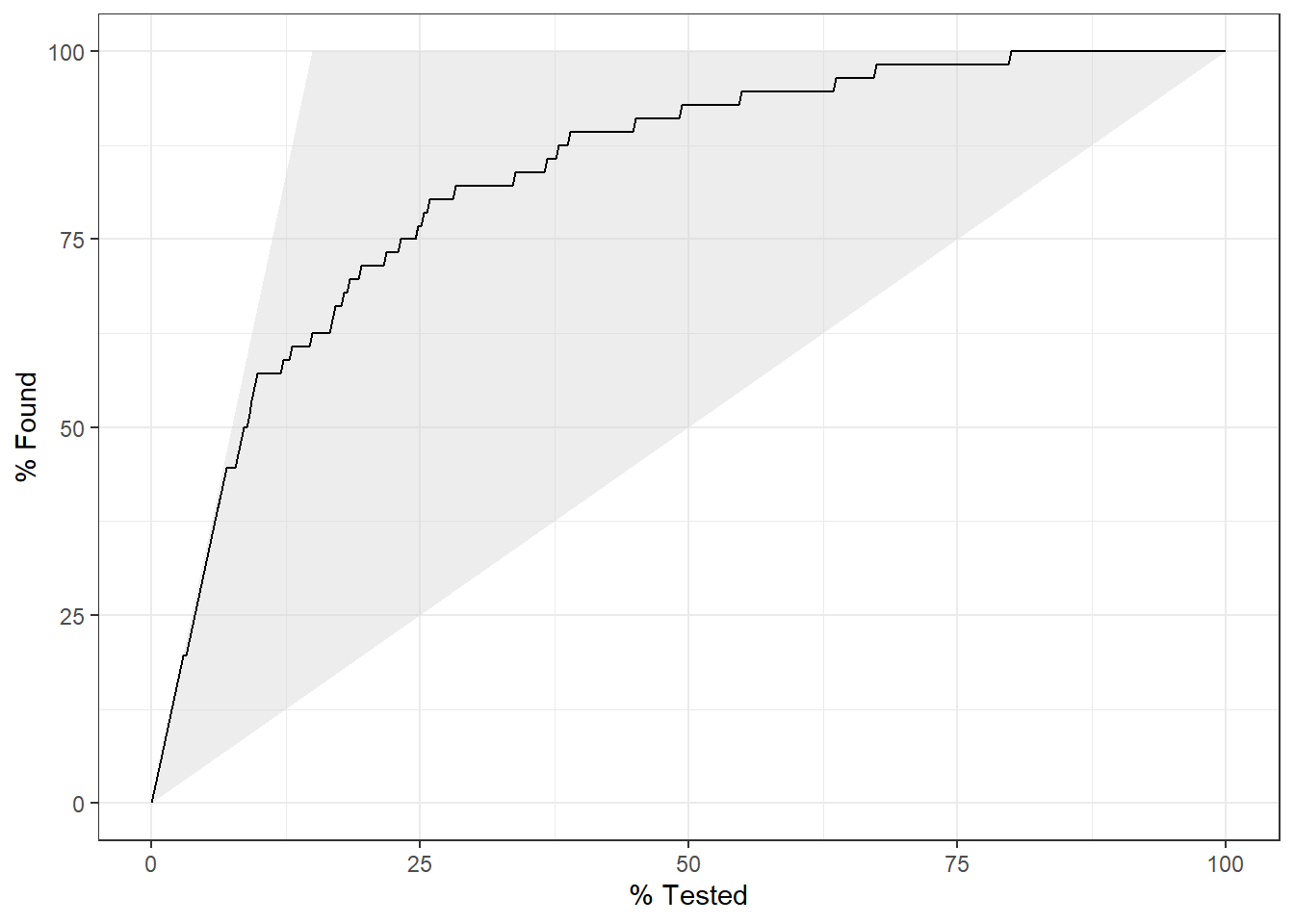
test.scored %>%
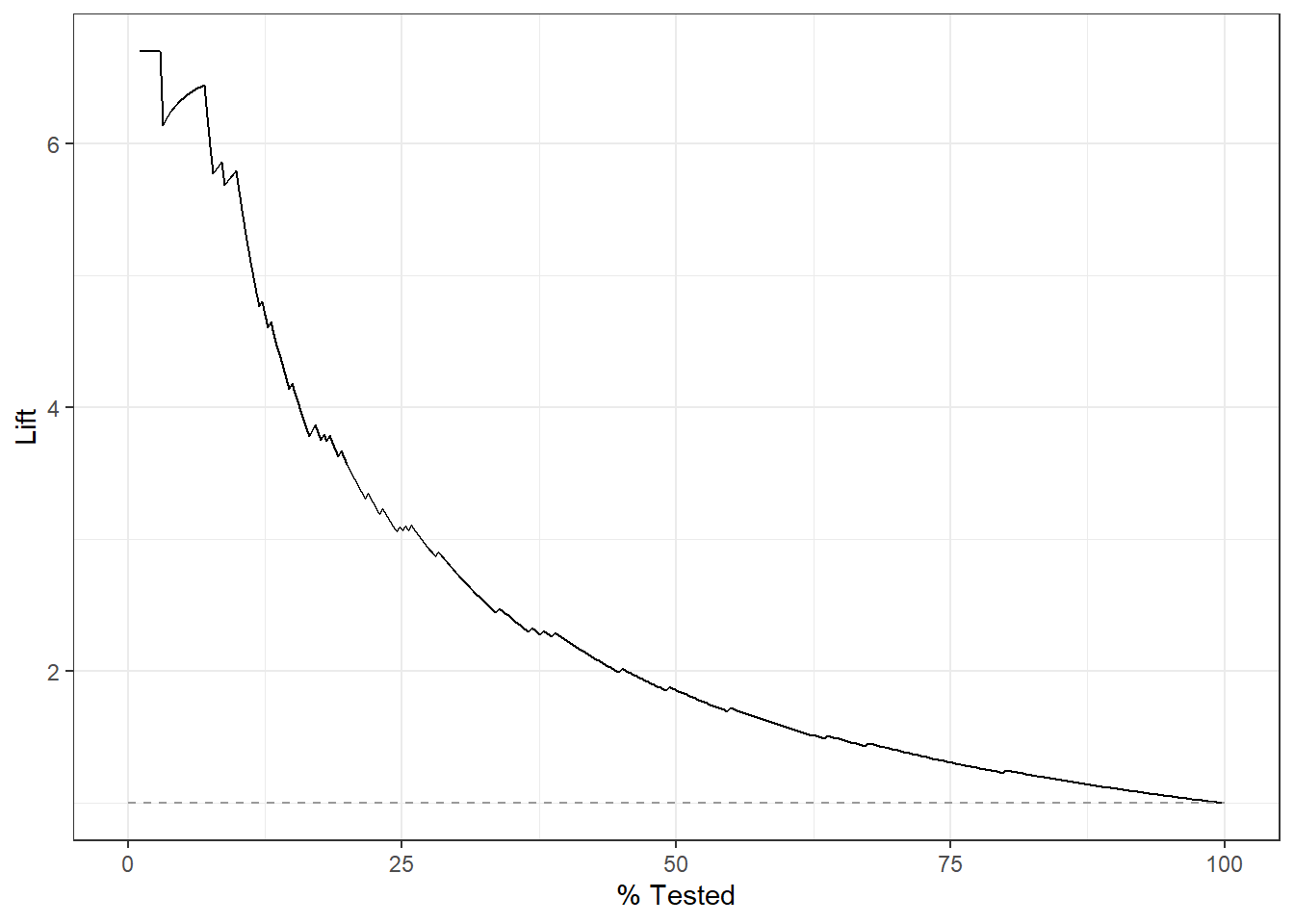
lift_curve(truth=diq010, Diabetes) %>%
autoplot()