── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors25 Random Forest Imputation and Multi-class Classifier
\(~\)
\(~\)
25.1 NHANES data
25.1.1 Note that Depressed is has 3 potiential classes
# A tibble: 3 × 1
Depressed
<fct>
1 Several
2 None
3 Most \(~\)
\(~\)
\(~\)
\(~\)
25.2 Some data prep
SumNa <- function(col){sum(is.na(col))}
data.sum <- NHANES_DATA_12 %>%
summarise_all(SumNa) %>%
tidyr::gather(key='feature', value='SumNa') %>%
arrange(-SumNa) %>%
mutate(PctNa = SumNa/nrow(NHANES_DATA_12))
data.sum2 <- data.sum %>%
filter(! (feature %in% c('ID','Depressed'))) %>%
filter(PctNa < .85)
data.sum2$feature [1] "UrineFlow2" "UrineVol2" "AgeRegMarij" "PregnantNow"
[5] "Age1stBaby" "nBabies" "nPregnancies" "SmokeAge"
[9] "AgeFirstMarij" "SmokeNow" "Testosterone" "AgeMonths"
[13] "TVHrsDay" "Race3" "CompHrsDay" "PhysActiveDays"
[17] "SexOrientation" "AlcoholDay" "SexNumPartYear" "Marijuana"
[21] "RegularMarij" "SexAge" "SexNumPartnLife" "HardDrugs"
[25] "SexEver" "SameSex" "AlcoholYear" "HHIncome"
[29] "HHIncomeMid" "Poverty" "UrineFlow1" "BPSys1"
[33] "BPDia1" "AgeDecade" "DirectChol" "TotChol"
[37] "BPSys2" "BPDia2" "Education" "BPSys3"
[41] "BPDia3" "MaritalStatus" "Smoke100" "Smoke100n"
[45] "Alcohol12PlusYr" "BPSysAve" "BPDiaAve" "Pulse"
[49] "BMI_WHO" "BMI" "Weight" "HomeRooms"
[53] "Height" "HomeOwn" "UrineVol1" "SleepHrsNight"
[57] "LittleInterest" "DaysPhysHlthBad" "DaysMentHlthBad" "Diabetes"
[61] "Work" "SurveyYr" "Gender" "Age"
[65] "Race1" "HealthGen" "SleepTrouble" "PhysActive"
25.2.1 note that data_F still has missing values
Amelia::missmap(as.data.frame(data_F))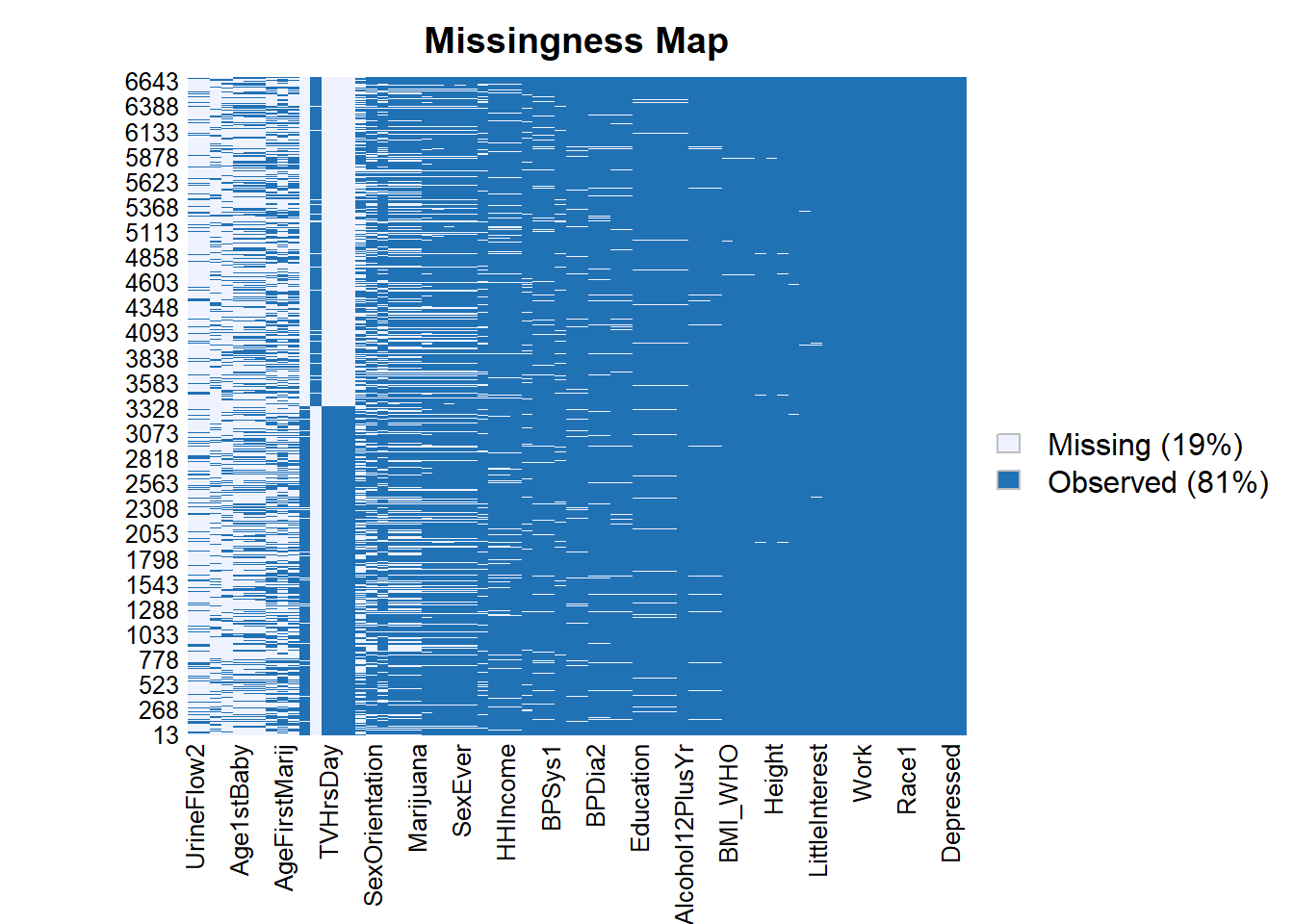
\(~\)
\(~\)
\(~\)
\(~\)
25.3 Random Forest Impute with rfImpute
randomForest 4.7-1.1Type rfNews() to see new features/changes/bug fixes.
Attaching package: 'randomForest'The following object is masked from 'package:dplyr':
combineThe following object is masked from 'package:ggplot2':
margindata_F.imputed <- rfImpute(Depressed ~ . ,
data_F,
iter=2,
ntree=300)ntree OOB 1 2 3
300: 8.08% 0.82% 36.57% 30.38%
ntree OOB 1 2 3
300: 8.98% 1.20% 38.26% 35.89%25.3.0.1 Note we no longer have missing data
Amelia::missmap(as.data.frame(data_F.imputed))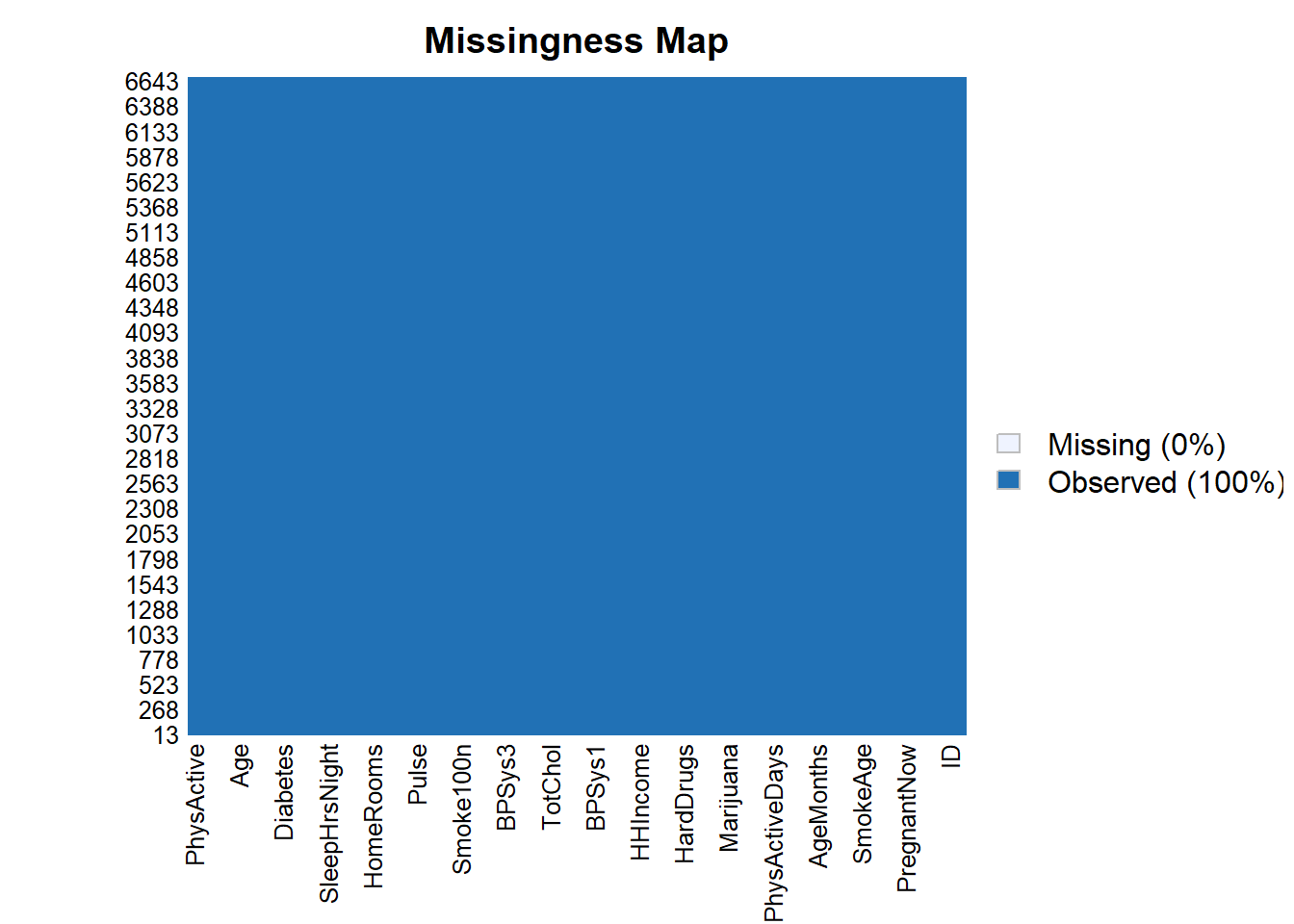
\(~\)
\(~\)
\(~\)
\(~\)
25.4 Split Data
Loading required package: lattice
Attaching package: 'caret'The following object is masked from 'package:purrr':
liftset.seed(8576309)
trainIndex <- createDataPartition(data_F.imputed$Depressed,
p = .6,
list = FALSE,
times = 1)
TRAIN <- data_F.imputed[trainIndex, ]
TEST <- data_F.imputed[-trainIndex, ]\(~\)
\(~\)
\(~\)
\(~\)
25.5 Train model
train_ctrl <- trainControl(method="cv", # type of resampling in this case Cross-Validated
number=3, # number of folds
search = "random", # we are performing a "random
)
toc <- Sys.time()
model_rf <- train(Depressed ~ .,
data = TRAIN,
method = "rf", # this will use the randomForest::randomForest function
metric = "Accuracy", # which metric should be optimized for
trControl = train_ctrl,
# options to be passed to randomForest
ntree = 741,
keep.forest=TRUE,
importance=TRUE)
tic <- Sys.time()
tic - tocTime difference of 3.258477 mins25.5.1 Model output
model_rfRandom Forest
4005 samples
69 predictor
3 classes: 'None', 'Several', 'Most'
No pre-processing
Resampling: Cross-Validated (3 fold)
Summary of sample sizes: 2670, 2669, 2671
Resampling results across tuning parameters:
mtry Accuracy Kappa
11 0.8781526 0.5935030
63 0.8846449 0.6435894
75 0.8853945 0.6477631
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was mtry = 75.randomForest::varImpPlot(model_rf$finalModel)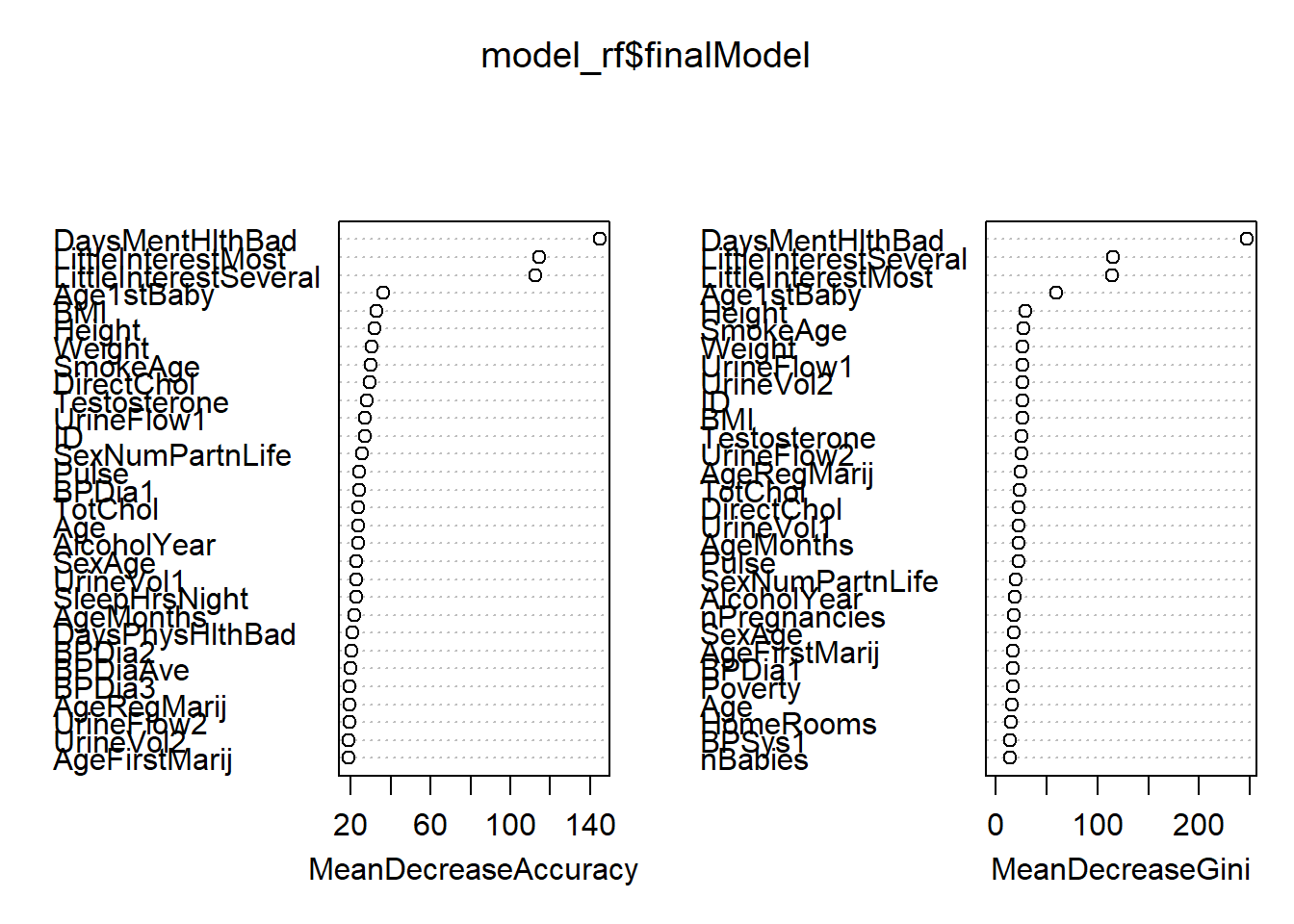
\(~\)
\(~\)
\(~\)
\(~\)
25.6 Score Test Data
25.6.1 Use yardstick for Model Metrics
Attaching package: 'yardstick'The following objects are masked from 'package:caret':
precision, recall, sensitivity, specificityThe following object is masked from 'package:readr':
speccm <- conf_mat(TEST.scored, truth = Depressed, class)
cm Truth
Prediction None Several Most
None 2030 160 24
Several 57 215 23
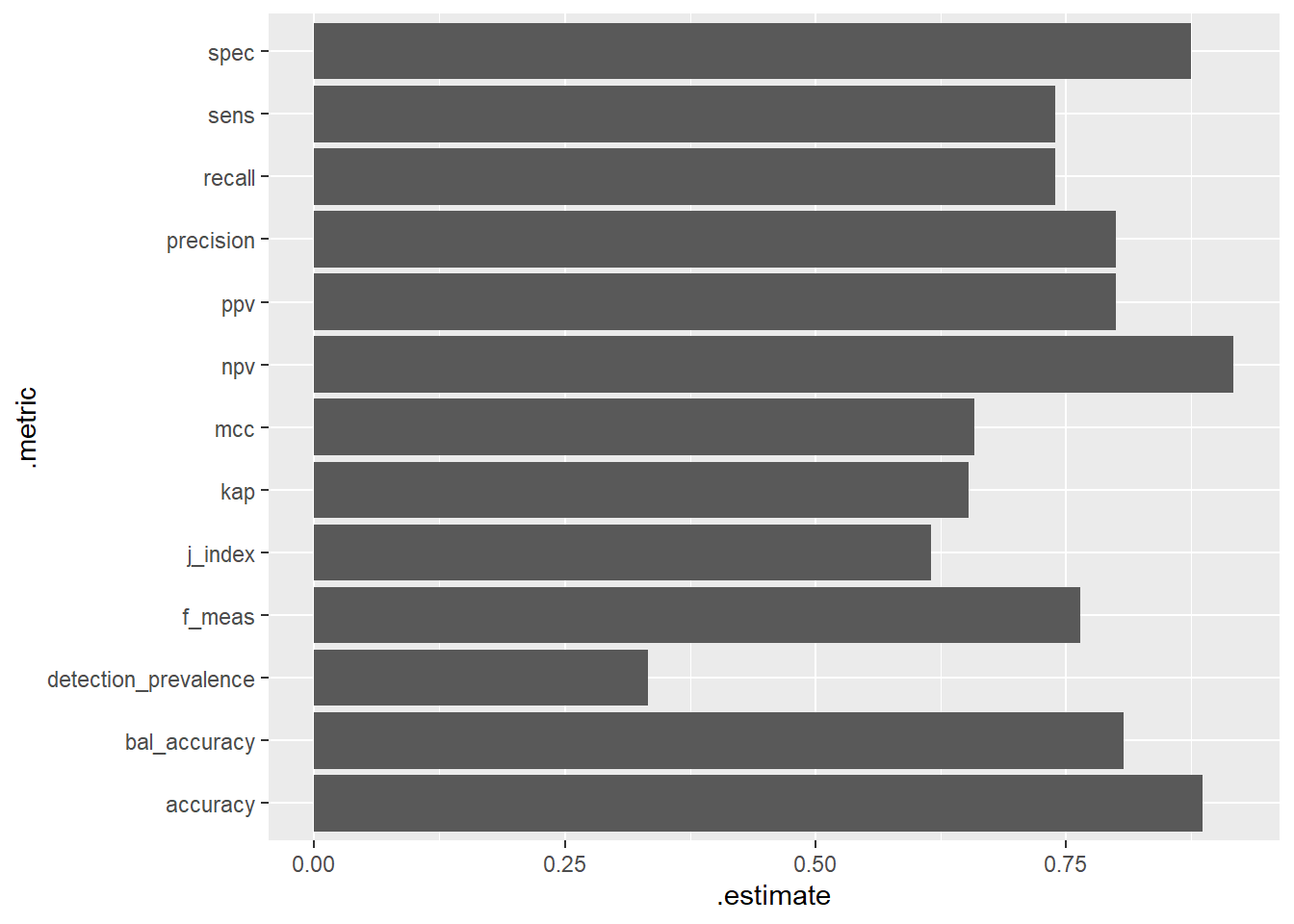
Most 11 28 120summary(cm)# A tibble: 13 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 accuracy multiclass 0.886
2 kap multiclass 0.653
3 sens macro 0.740
4 spec macro 0.875
5 ppv macro 0.800
6 npv macro 0.917
7 mcc multiclass 0.659
8 j_index macro 0.615
9 bal_accuracy macro 0.808
10 detection_prevalence macro 0.333
11 precision macro 0.800
12 recall macro 0.740
13 f_meas macro 0.765library('ggplot2')
ggplot(summary(cm), aes(x=.metric, y=.estimate)) +
geom_bar(stat="identity") +
coord_flip()