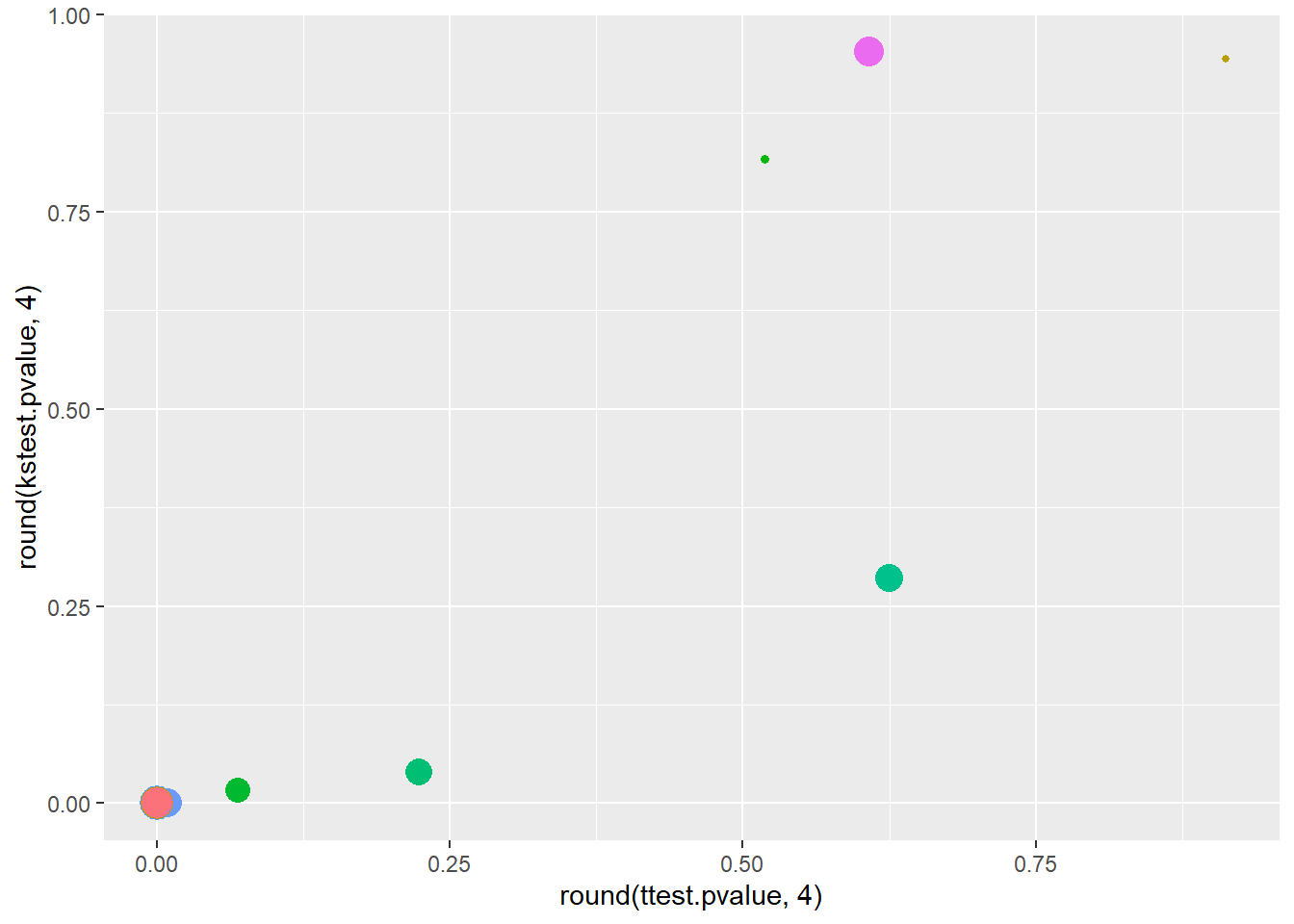
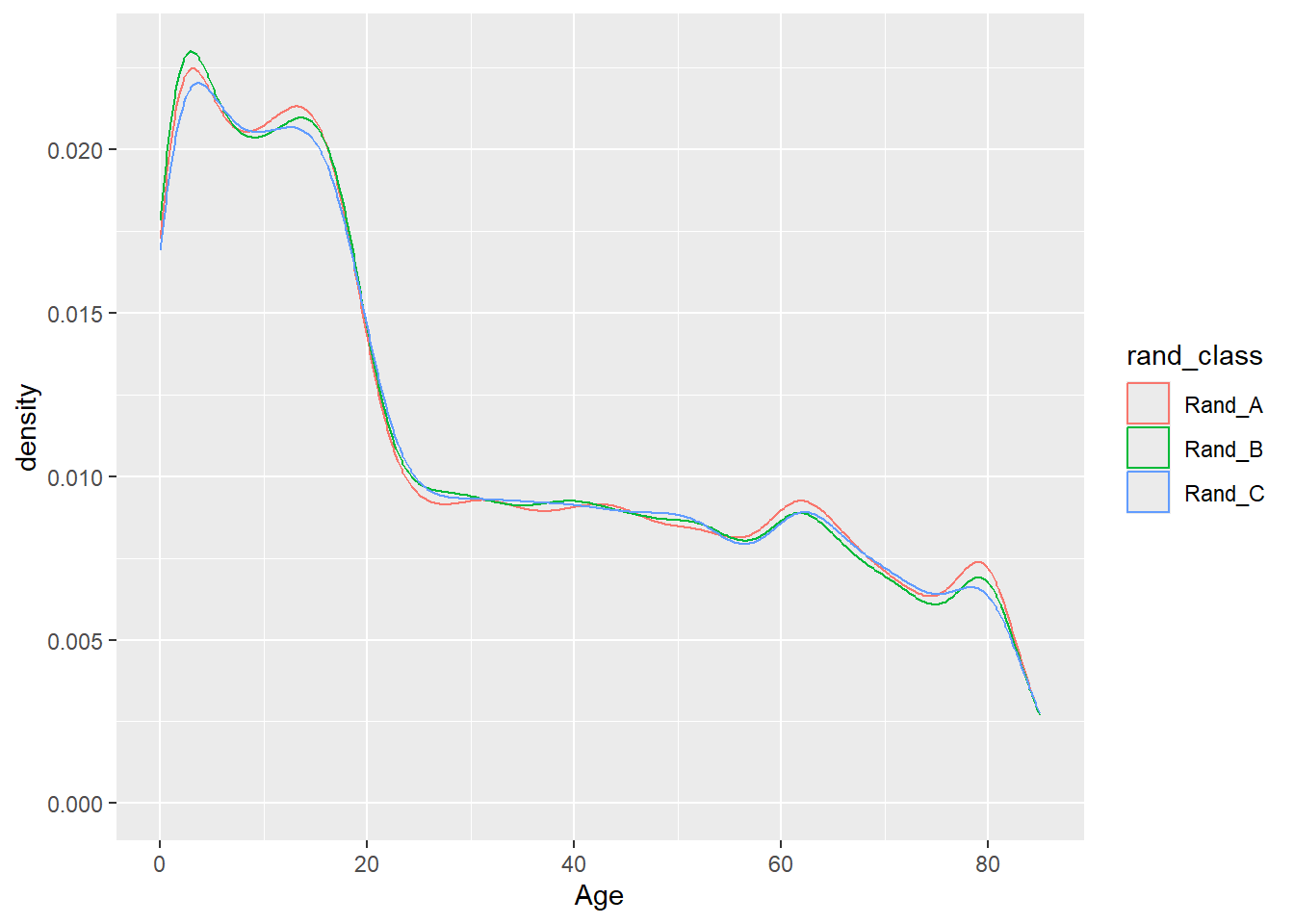
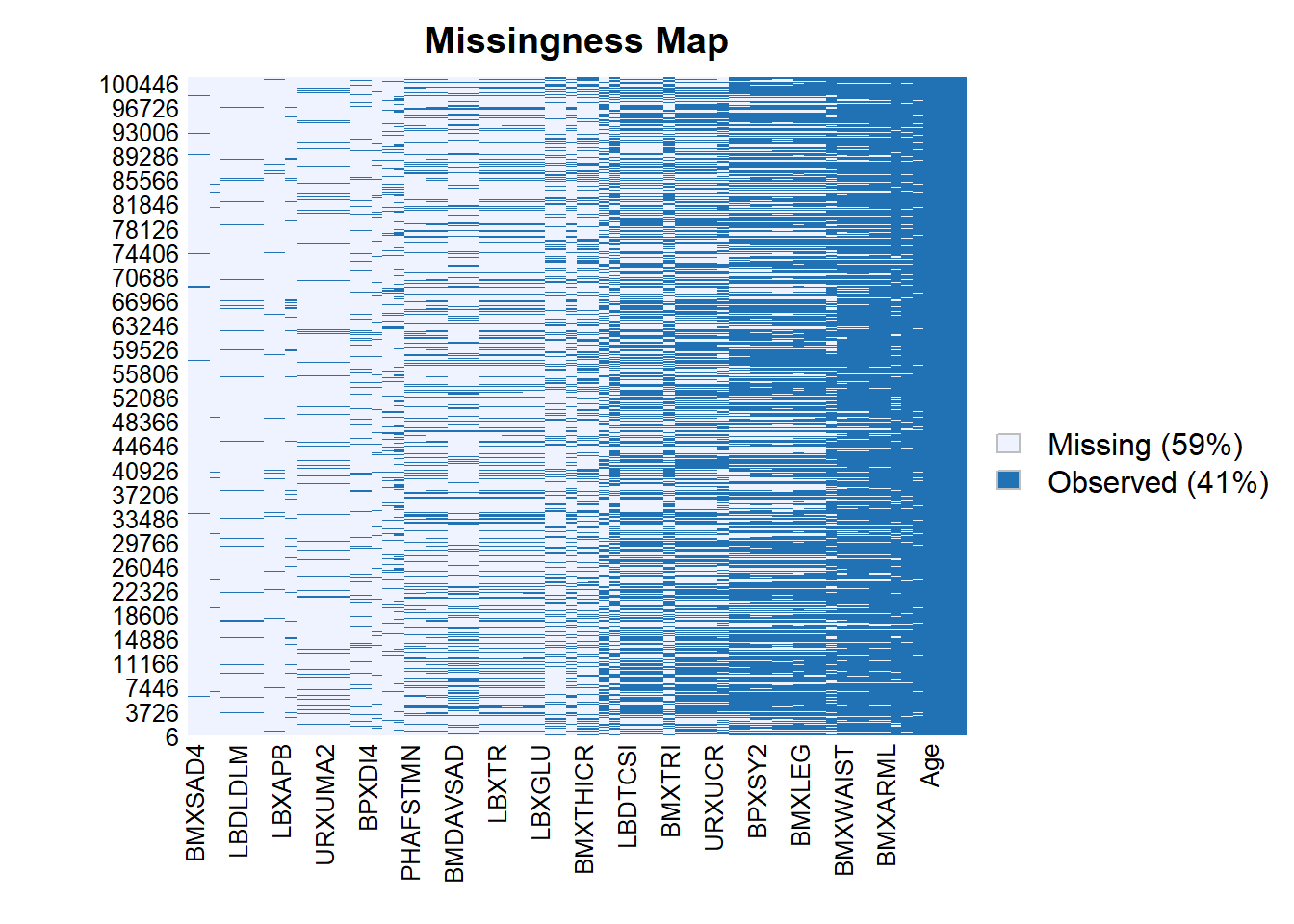
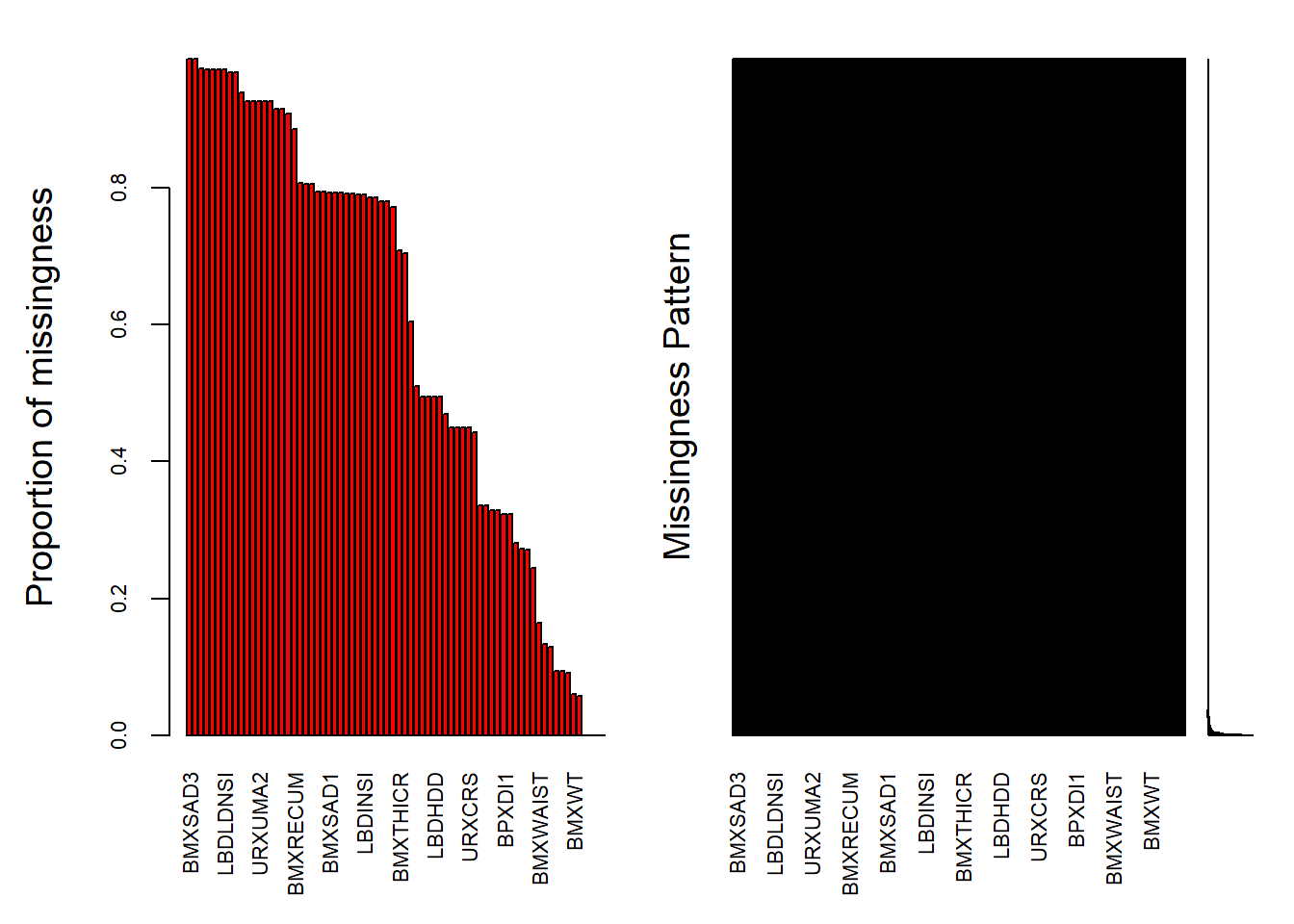
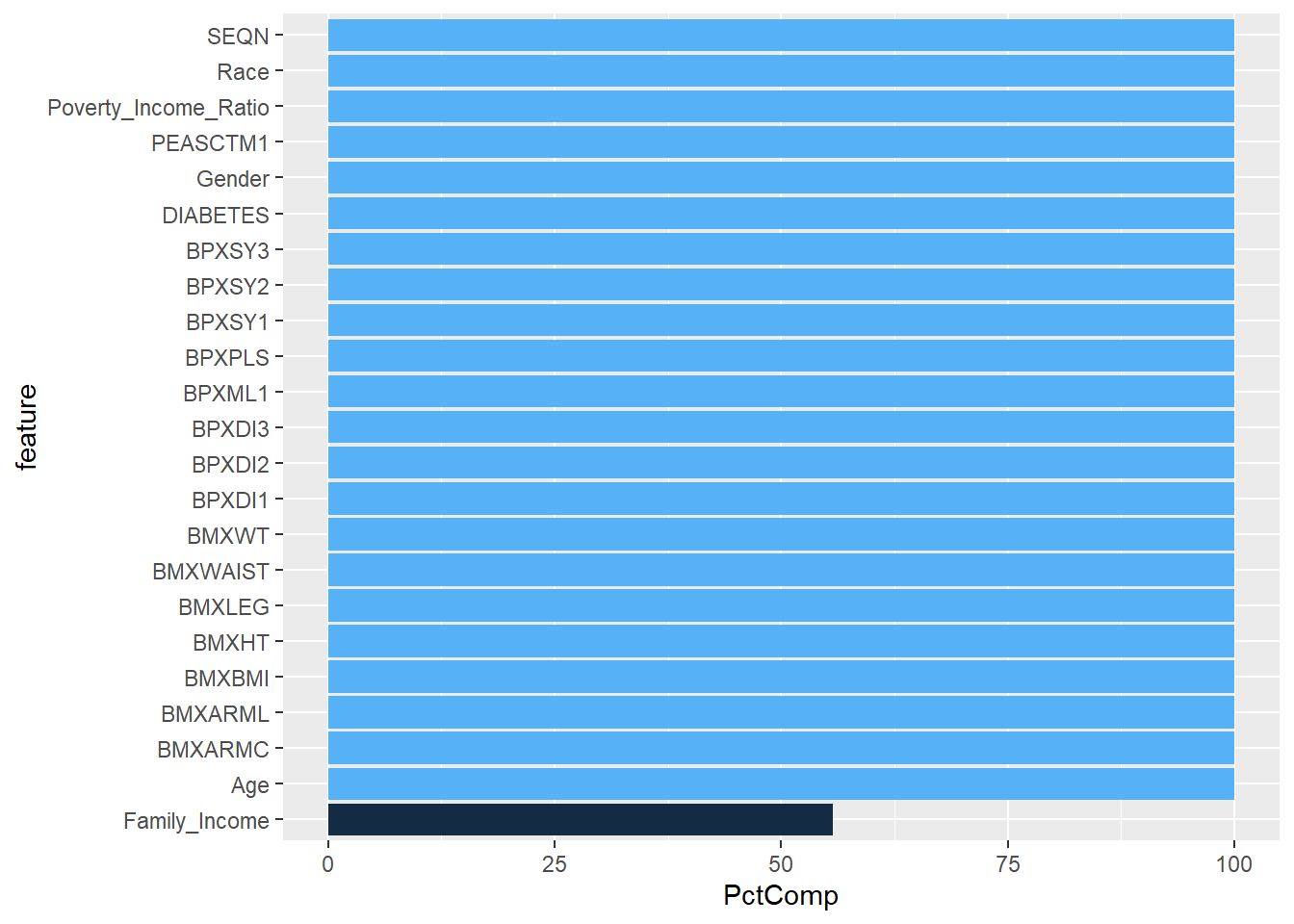
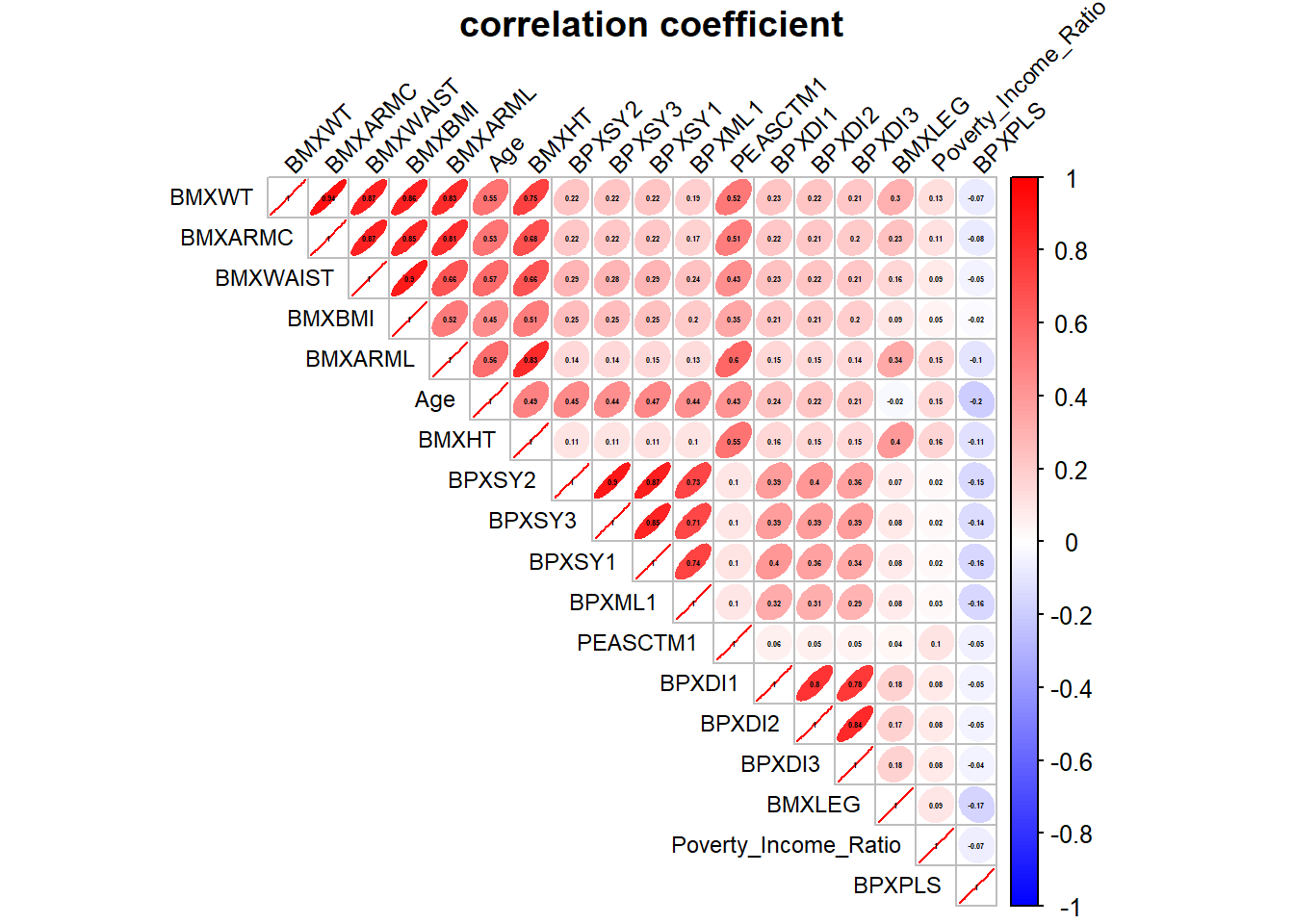
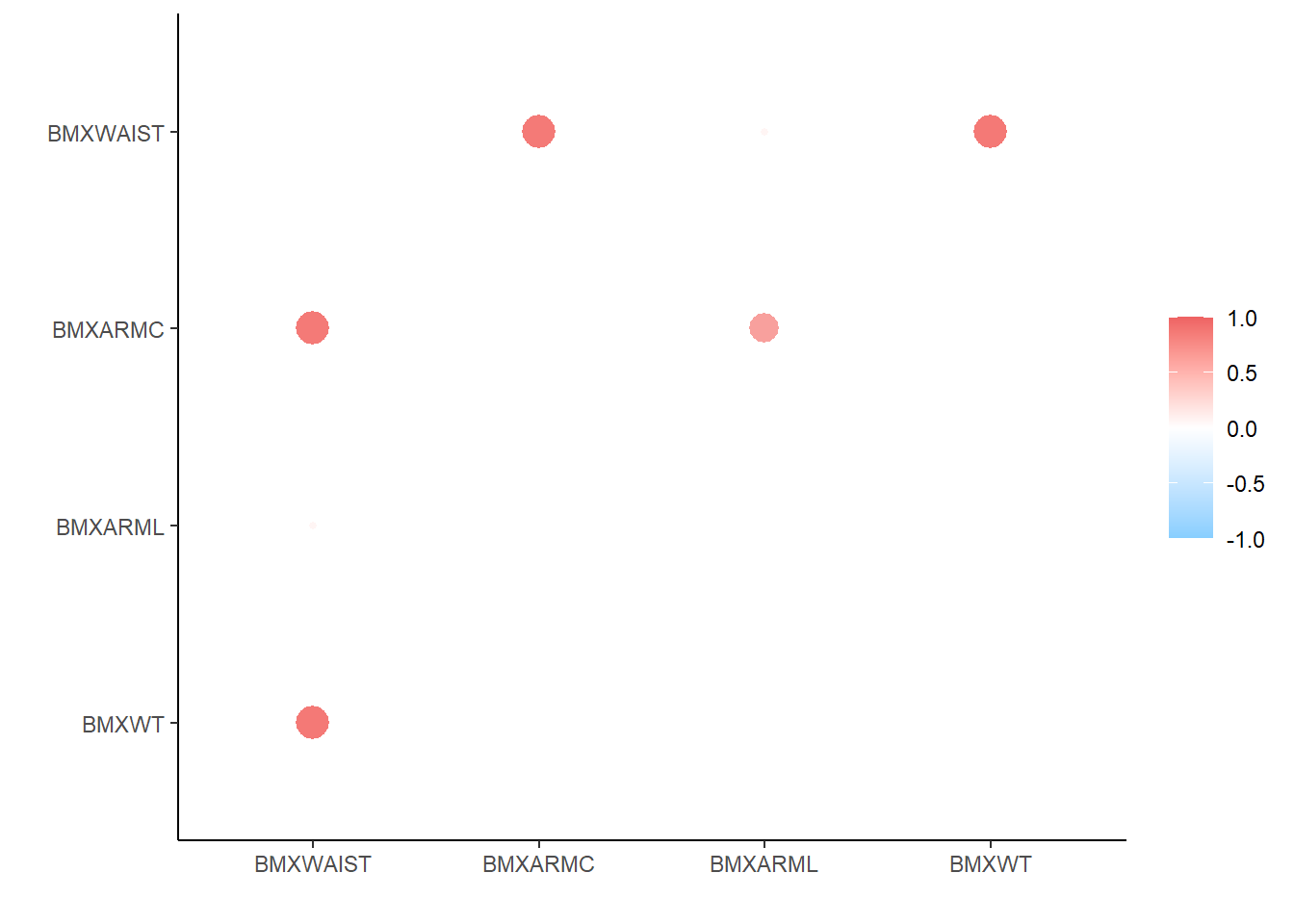
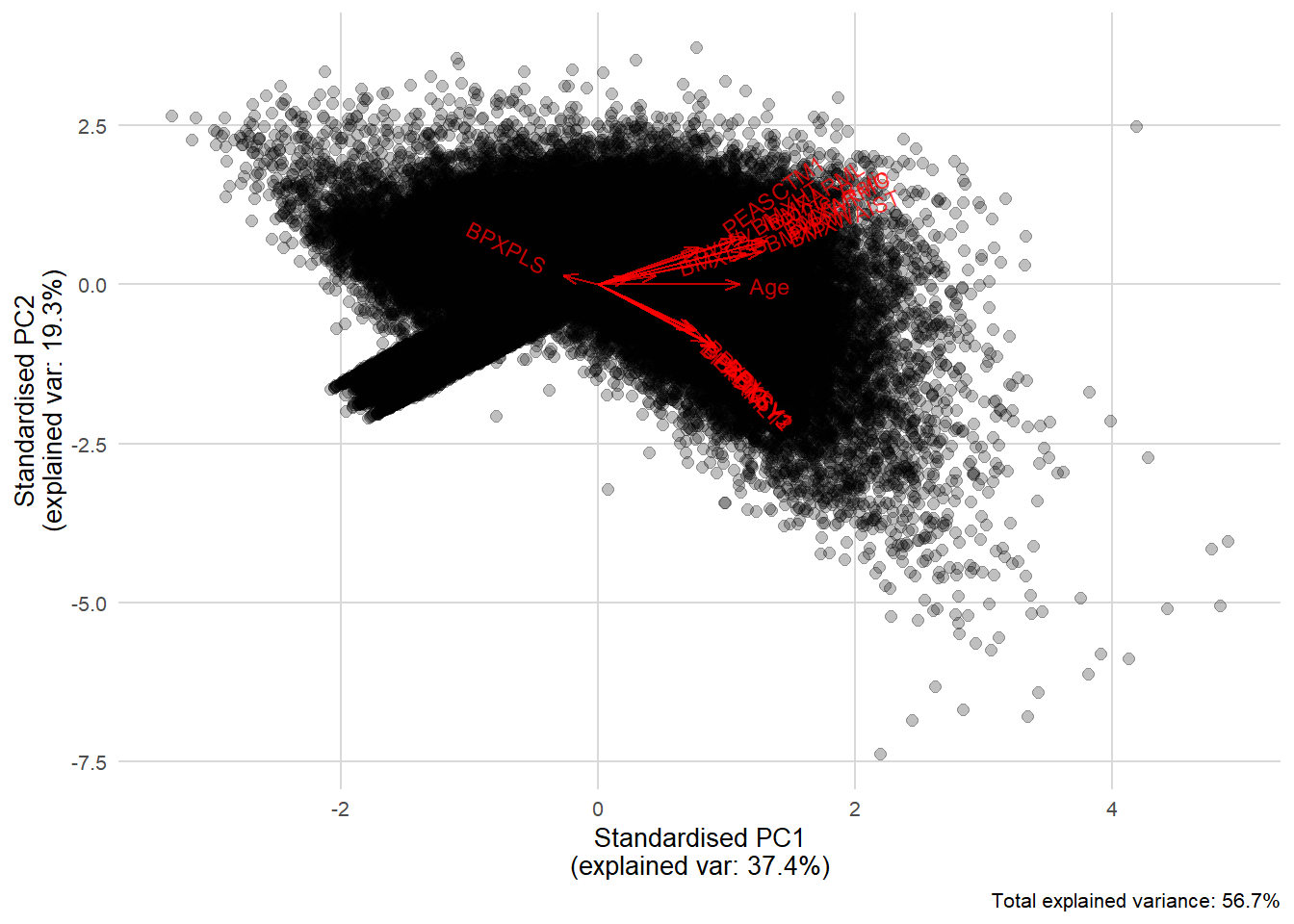
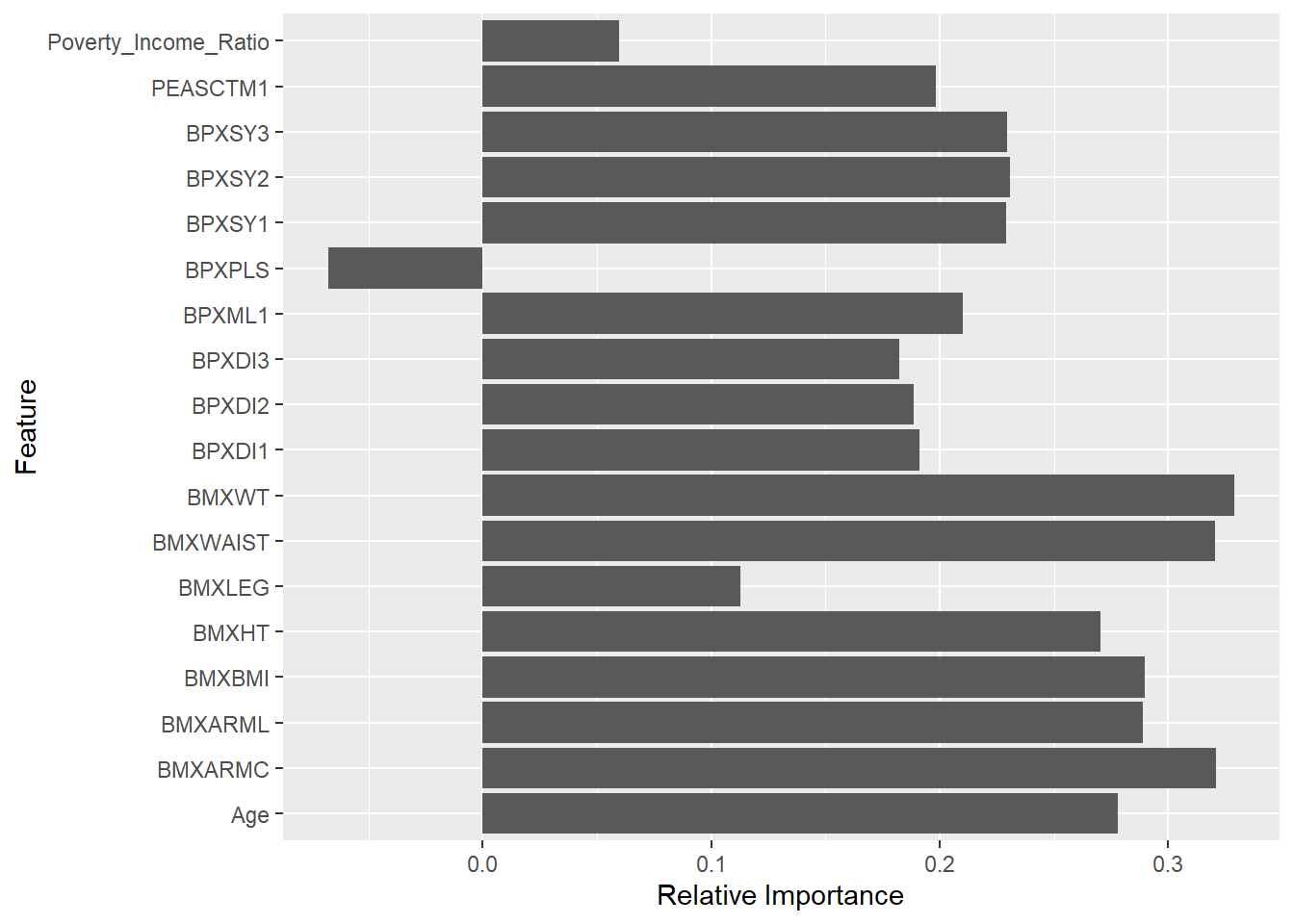
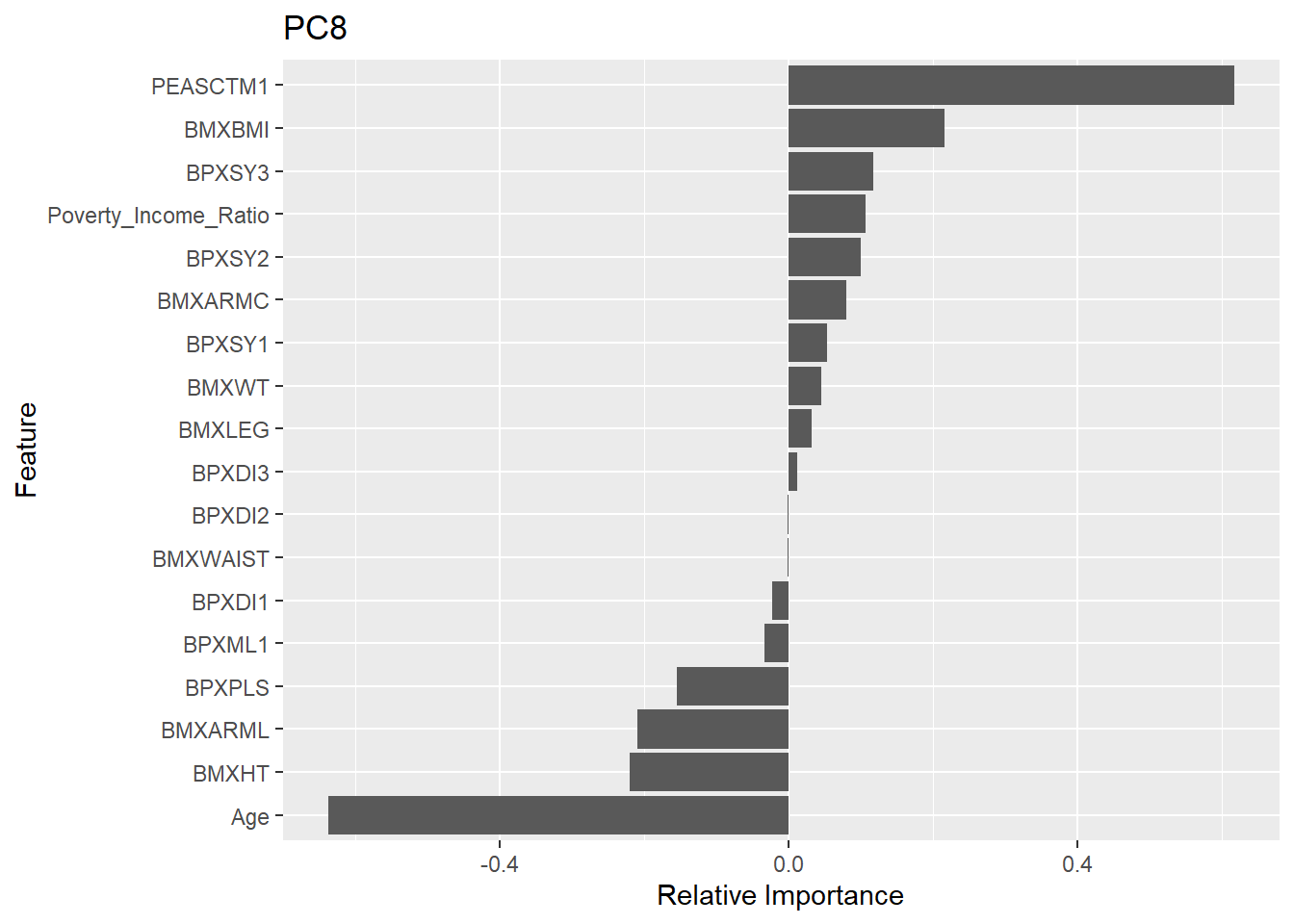
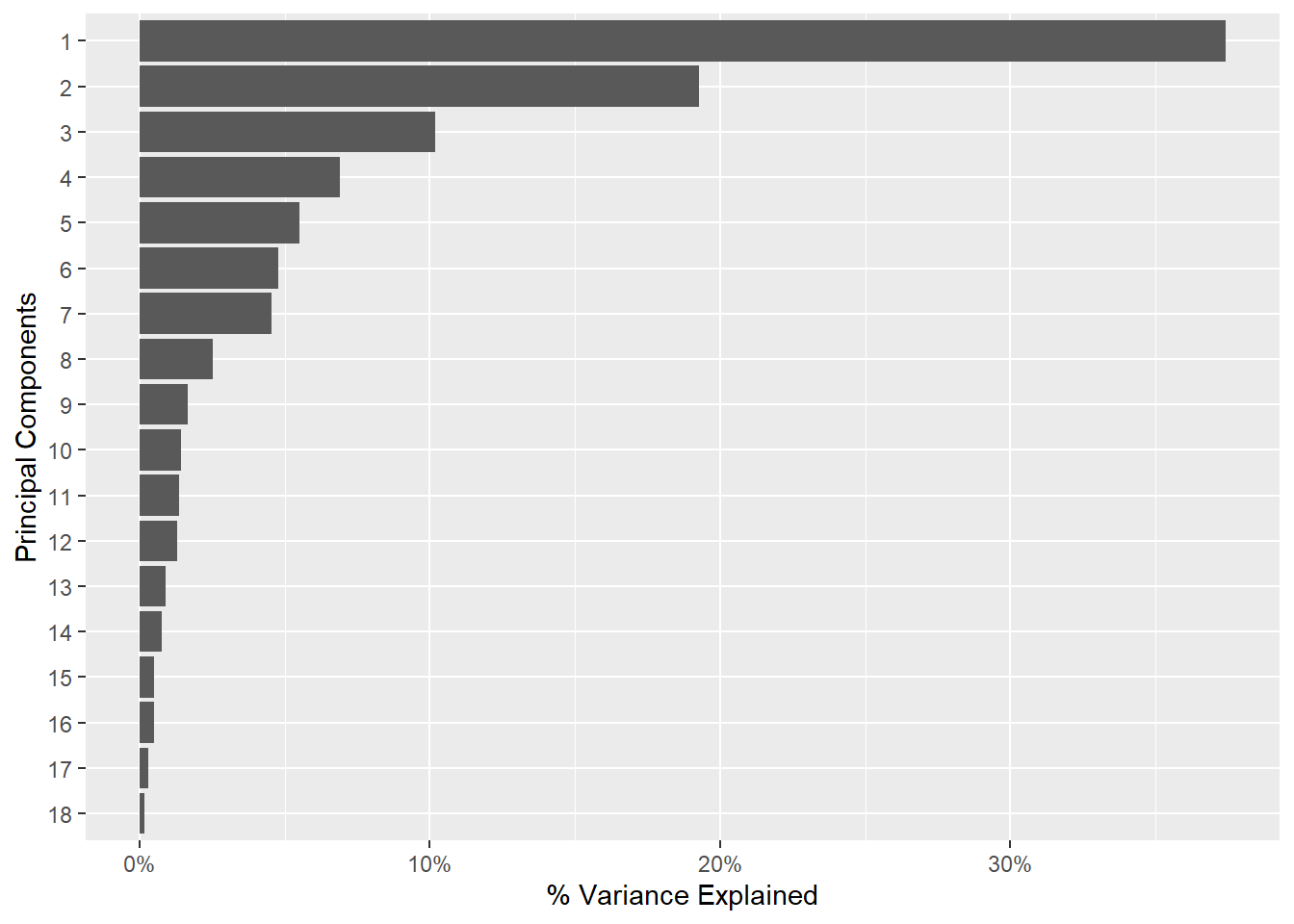
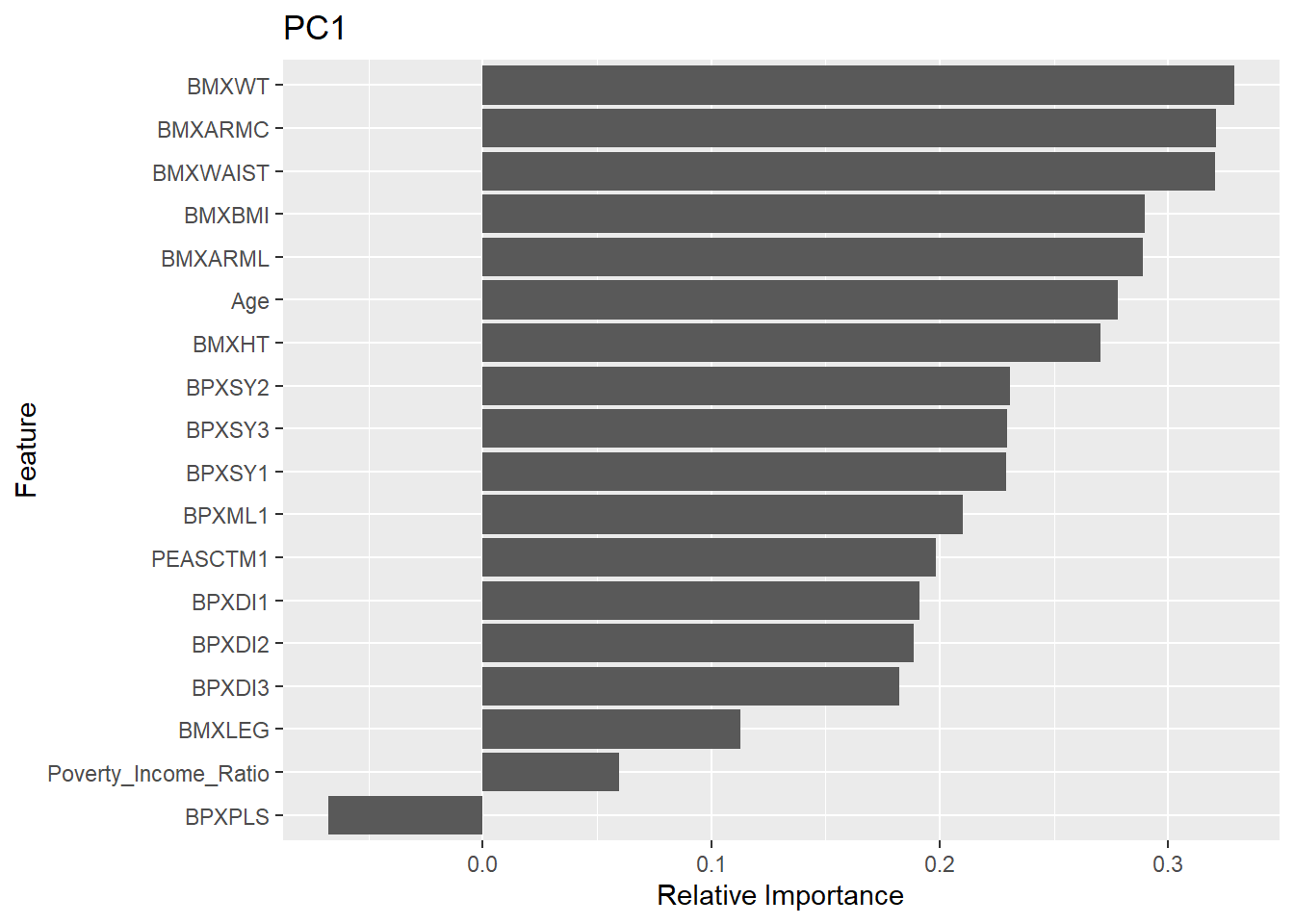
Rows: 150
Columns: 129
Groups: Gender, Race, Family_Income [150]
$ Gender <chr> "Female", "Female", "Female", "Female", "F…
$ Race <chr> "Black", "Black", "Black", "Black", "Black…
$ Family_Income <chr> "$ 0 to $ 4,999", "$ 5,000 to $ 9,999", "$…
$ BPXDI3_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXDI3_n_miss <int> 110, 142, 155, 117, 136, 36, 134, 206, 138…
$ BPXDI3_min <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 26,…
$ BPXDI3_max <dbl> 116, 126, 112, 110, 108, 102, 112, 118, 11…
$ BPXDI3_mean <dbl> 65.09449, 67.35016, 64.74659, 65.93396, 66…
$ BPXDI3_median <dbl> 66, 68, 66, 68, 68, 70, 66, 68, 66, 68, 68…
$ BPXDI3_sd <dbl> 16.55506, 17.14552, 16.63621, 15.14111, 16…
$ BPXSY3_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXSY3_n_miss <int> 110, 142, 155, 117, 136, 36, 134, 206, 138…
$ BPXSY3_min <dbl> 84, 82, 74, 82, 86, 80, 82, 80, 78, 82, 86…
$ BPXSY3_max <dbl> 224, 212, 218, 210, 196, 222, 212, 232, 20…
$ BPXSY3_mean <dbl> 117.7087, 123.9558, 122.2616, 116.7594, 12…
$ BPXSY3_median <dbl> 114, 118, 116, 112, 116, 122, 114, 116, 11…
$ BPXSY3_sd <dbl> 19.76127, 23.39178, 23.76564, 18.70560, 20…
$ BPXDI2_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXDI2_n_miss <int> 114, 145, 160, 112, 129, 34, 126, 208, 138…
$ BPXDI2_min <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 36,…
$ BPXDI2_max <dbl> 126, 124, 110, 114, 110, 104, 108, 122, 11…
$ BPXDI2_mean <dbl> 65.25600, 67.51592, 65.16575, 66.54079, 66…
$ BPXDI2_median <dbl> 66, 66, 66, 68, 68, 68, 66, 66, 66, 66, 66…
$ BPXDI2_sd <dbl> 15.90050, 16.45426, 16.46419, 14.67869, 16…
$ BPXSY2_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXSY2_n_miss <int> 114, 145, 160, 112, 129, 34, 126, 208, 138…
$ BPXSY2_min <dbl> 86, 86, 72, 80, 86, 84, 84, 84, 76, 82, 86…
$ BPXSY2_max <dbl> 190, 228, 212, 208, 194, 216, 210, 234, 20…
$ BPXSY2_mean <dbl> 117.6400, 125.1401, 123.3702, 117.6364, 12…
$ BPXSY2_median <dbl> 114, 120, 116, 114, 116, 122, 114, 116, 11…
$ BPXSY2_sd <dbl> 19.00957, 23.64783, 23.73587, 18.99184, 21…
$ BPXDI1_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXDI1_n_miss <int> 113, 148, 163, 129, 139, 38, 138, 219, 145…
$ BPXDI1_min <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 42,…
$ BPXDI1_max <dbl> 124, 124, 106, 114, 104, 112, 116, 116, 11…
$ BPXDI1_mean <dbl> 66.28685, 68.47588, 65.14206, 66.51942, 66…
$ BPXDI1_median <dbl> 66, 68, 66, 68, 68, 68, 66, 66, 66, 68, 68…
$ BPXDI1_sd <dbl> 14.07457, 15.88685, 15.49407, 13.23757, 15…
$ BPXSY1_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXSY1_n_miss <int> 113, 148, 163, 129, 139, 38, 138, 219, 145…
$ BPXSY1_min <dbl> 88, 80, 74, 82, 84, 84, 86, 84, 78, 84, 86…
$ BPXSY1_max <dbl> 208, 230, 220, 208, 198, 216, 222, 238, 20…
$ BPXSY1_mean <dbl> 117.8725, 125.1447, 122.8969, 117.5485, 12…
$ BPXSY1_median <dbl> 114, 120, 118, 114, 116, 122, 114, 116, 11…
$ BPXSY1_sd <dbl> 19.40844, 24.38247, 23.34555, 18.58550, 21…
$ BMXLEG_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXLEG_n_miss <int> 116, 139, 165, 111, 134, 35, 126, 199, 129…
$ BMXLEG_min <dbl> 27.6, 27.7, 24.9, 27.1, 27.2, 29.8, 28.0, …
$ BMXLEG_max <dbl> 47.2, 46.8, 48.2, 48.5, 50.0, 45.0, 46.7, …
$ BMXLEG_mean <dbl> 38.63629, 38.14375, 37.86022, 38.96349, 37…
$ BMXLEG_median <dbl> 38.95, 38.20, 38.00, 39.05, 38.00, 38.30, …
$ BMXLEG_sd <dbl> 3.544325, 3.598819, 3.773591, 3.155392, 3.…
$ BPXML1_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXML1_n_miss <int> 101, 130, 143, 104, 119, 31, 113, 184, 126…
$ BPXML1_min <dbl> 110, 0, 110, 100, 110, 120, 110, 110, 110,…
$ BPXML1_max <dbl> 240, 250, 888, 220, 220, 250, 888, 888, 24…
$ BPXML1_mean <dbl> 143.3840, 149.3617, 155.3879, 143.5515, 14…
$ BPXML1_median <dbl> 140, 140, 140, 140, 140, 140, 140, 140, 14…
$ BPXML1_sd <dbl> 20.31134, 24.59216, 69.76214, 18.14822, 21…
$ BPXPLS_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BPXPLS_n_miss <int> 101, 131, 142, 104, 119, 31, 113, 183, 126…
$ BPXPLS_min <dbl> 50, 46, 46, 44, 46, 50, 46, 46, 50, 48, 36…
$ BPXPLS_max <dbl> 120, 116, 130, 120, 112, 110, 140, 118, 12…
$ BPXPLS_mean <dbl> 77.87072, 76.92073, 75.43684, 74.52174, 75…
$ BPXPLS_median <dbl> 78, 76, 74, 74, 74, 74, 76, 76, 74, 74, 74…
$ BPXPLS_sd <dbl> 12.51924, 12.55665, 12.28718, 11.47804, 12…
$ PEASCTM1_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ PEASCTM1_n_miss <int> 131, 145, 170, 229, 155, 48, 187, 294, 229…
$ PEASCTM1_min <dbl> 6, 9, 6, 37, 7, 46, 8, 3, 7, 6, 5, 7, 6, 1…
$ PEASCTM1_max <dbl> 1131, 1536, 1243, 1439, 1274, 1086, 1142, …
$ PEASCTM1_mean <dbl> 543.7597, 563.0000, 554.6705, 637.6538, 55…
$ PEASCTM1_median <dbl> 567.0, 576.5, 603.5, 657.5, 590.0, 667.0, …
$ PEASCTM1_sd <dbl> 248.5154, 272.9288, 271.9278, 245.9812, 25…
$ BMXWAIST_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXWAIST_n_miss <int> 59, 66, 79, 66, 55, 25, 73, 91, 55, 57, 26…
$ BMXWAIST_min <dbl> 38.7, 43.3, 40.5, 46.2, 42.7, 43.9, 44.2, …
$ BMXWAIST_max <dbl> 165.5, 171.6, 156.8, 163.5, 158.8, 151.7, …
$ BMXWAIST_mean <dbl> 83.58656, 89.62290, 88.33115, 89.26379, 84…
$ BMXWAIST_median <dbl> 80.80, 90.20, 91.60, 89.40, 86.70, 99.60, …
$ BMXWAIST_sd <dbl> 25.32800, 26.69575, 25.81373, 21.50595, 24…
$ BMXBMI_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXBMI_n_miss <int> 37, 36, 45, 42, 32, 15, 39, 59, 36, 32, 11…
$ BMXBMI_min <dbl> 13.40, 12.50, 13.41, 13.40, 12.70, 13.30, …
$ BMXBMI_max <dbl> 57.80, 82.10, 77.50, 68.70, 84.40, 64.70, …
$ BMXBMI_mean <dbl> 26.12003, 27.81638, 27.77199, 27.98603, 26…
$ BMXBMI_median <dbl> 23.700, 26.660, 27.230, 26.900, 25.500, 30…
$ BMXBMI_sd <dbl> 9.509575, 10.418529, 9.889805, 8.842373, 9…
$ BMXHT_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXHT_n_miss <int> 36, 36, 44, 42, 32, 14, 39, 59, 34, 32, 11…
$ BMXHT_min <dbl> 78.5, 81.6, 82.8, 86.0, 82.5, 92.3, 86.9, …
$ BMXHT_max <dbl> 184.8, 186.4, 180.9, 187.8, 184.1, 177.4, …
$ BMXHT_mean <dbl> 148.9064, 150.9000, 150.5358, 156.7822, 15…
$ BMXHT_median <dbl> 158.45, 159.00, 158.35, 161.40, 158.50, 16…
$ BMXHT_sd <dbl> 23.88131, 22.45114, 21.96616, 16.70671, 22…
$ BMXARMC_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXARMC_n_miss <int> 41, 40, 58, 44, 41, 20, 50, 65, 39, 42, 20…
$ BMXARMC_min <dbl> 13.8, 13.8, 13.9, 14.6, 13.7, 14.0, 13.1, …
$ BMXARMC_max <dbl> 54.0, 57.3, 51.7, 58.1, 48.5, 51.5, 58.3, …
$ BMXARMC_mean <dbl> 28.14458, 29.31217, 29.28405, 30.31288, 28…
$ BMXARMC_median <dbl> 28.00, 29.80, 30.10, 30.70, 29.30, 32.50, …
$ BMXARMC_sd <dbl> 8.507530, 9.030110, 8.539857, 7.509531, 8.…
$ BMXARML_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXARML_n_miss <int> 43, 42, 57, 45, 40, 22, 52, 66, 39, 42, 21…
$ BMXARML_min <dbl> 13.9, 15.0, 14.4, 16.0, 14.0, 15.6, 16.0, …
$ BMXARML_max <dbl> 42.0, 43.9, 43.2, 43.0, 43.9, 41.9, 43.0, …
$ BMXARML_mean <dbl> 32.38505, 32.86547, 33.19505, 34.53145, 32…
$ BMXARML_median <dbl> 34.60, 35.40, 35.70, 36.00, 35.15, 36.00, …
$ BMXARML_sd <dbl> 6.895394, 6.810559, 6.819470, 5.132052, 6.…
$ Poverty_Income_Ratio_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ Poverty_Income_Ratio_n_miss <int> 0, 0, 0, 0, 0, 171, 0, 0, 0, 0, 0, 0, 0, 1…
$ Poverty_Income_Ratio_min <dbl> 0.00, 0.12, 0.27, 2.25, 0.30, Inf, 0.48, 0…
$ Poverty_Income_Ratio_max <dbl> 0.43, 0.95, 1.35, 5.00, 1.85, -Inf, 2.31, …
$ Poverty_Income_Ratio_mean <dbl> 0.10730769, 0.46331155, 0.70402299, 4.6876…
$ Poverty_Income_Ratio_median <dbl> 0.085, 0.410, 0.680, 5.000, 0.860, NA, 1.0…
$ Poverty_Income_Ratio_sd <dbl> 0.09773131, 0.20340318, 0.26553590, 0.5764…
$ BMXWT_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ BMXWT_n_miss <int> 24, 17, 30, 27, 22, 9, 23, 36, 21, 19, 6, …
$ BMXWT_min <dbl> 7.7, 8.9, 8.5, 10.0, 8.9, 10.0, 8.9, 8.2, …
$ BMXWT_max <dbl> 157.5, 187.7, 191.6, 193.7, 219.6, 173.4, …
$ BMXWT_mean <dbl> 60.84735, 65.91312, 66.12337, 70.02490, 63…
$ BMXWT_median <dbl> 59.65, 67.10, 70.05, 70.55, 64.95, 76.75, …
$ BMXWT_sd <dbl> 33.33993, 35.45615, 33.77805, 29.73478, 33…
$ Age_n <int> 364, 459, 522, 541, 456, 171, 544, 827, 57…
$ Age_n_miss <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ Age_min <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ Age_max <dbl> 80, 80, 80, 80, 80, 80, 80, 80, 80, 80, 80…
$ Age_mean <dbl> 24.66484, 33.91285, 35.53831, 33.82994, 31…
$ Age_median <dbl> 19.0, 28.0, 31.0, 35.0, 24.0, 42.0, 26.0, …
$ Age_sd <dbl> 20.24840, 25.68241, 26.35764, 22.24478, 25…